- Record: found
- Abstract: found
- Article: found
Hematopoietic stem cells and betaherpesvirus latency
Read this article at
Abstract
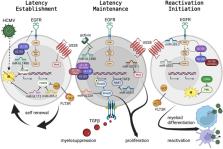
The human betaherpesviruses including human cytomegalovirus (HCMV), human herpesvirus (HHV)-6a and HHV-6b, and HHV-7 infect and establish latency in CD34+ hematopoietic stem and progenitor cells (HPCs). The diverse repertoire of HPCs in humans and the complex interactions between these viruses and host HPCs regulate the viral lifecycle, including latency. Precise manipulation of host and viral factors contribute to preferential maintenance of the viral genome, increased host cell survival, and specific manipulation of the cellular environment including suppression of neighboring cells and immune control. The dynamic control of these processes by the virus regulate inter- and intra-host signals critical to the establishment of chronic infection. Regulation occurs through direct viral protein interactions and cellular signaling, miRNA regulation, and viral mimics of cellular receptors and ligands, all leading to control of cell proliferation, survival, and differentiation. Hematopoietic stem cells have unique biological properties and the tandem control of virus and host make this a unique environment for chronic herpesvirus infection in the bone marrow. This review highlights the elegant complexities of the betaherpesvirus latency and HPC virus-host interactions.
Related collections
Most cited references187
- Record: found
- Abstract: found
- Article: not found
Proteomics. Tissue-based map of the human proteome.
- Record: found
- Abstract: found
- Article: not found
From haematopoietic stem cells to complex differentiation landscapes.
- Record: found
- Abstract: found
- Article: not found