- Record: found
- Abstract: found
- Article: found
High-level expression of the monomeric SARS-CoV-2 S protein RBD 320-537 in stably transfected CHO cells by the EEF1A1-based plasmid vector
Read this article at
Abstract
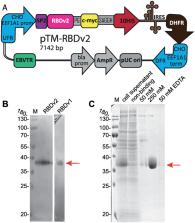
The spike (S) protein is one of the three proteins forming the coronaviruses’ viral envelope. The S protein of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has a spatial structure similar to the S proteins of other mammalian coronaviruses, except for a unique receptor-binding domain (RBD), which is a significant inducer of host immune response. Recombinant SARS-CoV-2 RBD is widely used as a highly specific minimal antigen for serological tests. Correct exposure of antigenic determinants has a significant impact on the accuracy of such tests–the antigen has to be correctly folded, contain no potentially antigenic non-vertebrate glycans, and, preferably, should have a glycosylation pattern similar to the native S protein. Based on the previously developed p1.1 vector, containing the regulatory sequences of the Eukaryotic translation elongation factor 1 alpha gene ( EEF1A1) from Chinese hamster, we created two expression constructs encoding SARS-CoV-2 RBD with C-terminal c-myc and polyhistidine tags. RBDv1 contained a native viral signal peptide, RBDv2 –human tPA signal peptide. We transfected a CHO DG44 cell line, selected stably transfected cells, and performed a few rounds of methotrexate-driven amplification of the genetic cassette in the genome. For the RBDv2 variant, a high-yield clonal producer cell line was obtained. We developed a simple purification scheme that consistently yielded up to 30 mg of RBD protein per liter of the simple shake flask cell culture. Purified proteins were analyzed by polyacrylamide gel electrophoresis in reducing and non-reducing conditions and gel filtration; for RBDv2 protein, the monomeric form content exceeded 90% for several series. Deglycosylation with PNGase F and mass spectrometry confirmed the presence of N-glycosylation. The antigen produced by the described technique is suitable for serological tests and subunit vaccine studies.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein
- Record: found
- Abstract: found
- Article: found
Structure, Function, and Evolution of Coronavirus Spike Proteins
- Record: found
- Abstract: found
- Article: not found