- Record: found
- Abstract: found
- Article: found
Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species
Read this article at
Significance
Natural products biosynthesized by cryptic gene clusters represent a largely untapped source for drug discovery. However, mining of these products by promoter engineering is restricted by the lack of streamlined genetic tools, especially in nonmodel biosynthetic gene cluster (BGC)-rich bacteria. Here, we describe the discovery of a pair of bacteriophage recombinases and application of recombinase-assisted promoter engineering to rapidly identify and activate several cryptic biosynthetic gene clusters in two Burkholderiales strains that currently lack effective genetic tools. Construction of an efficient genome engineering platform in a natural product producer expedites mining of cryptic BGCs in their native backgrounds, and host melioration for yield or structure optimization. This strategy enables potentially scalable discovery of novel metabolites with intriguing bioactivities from many other bacteria.
Abstract
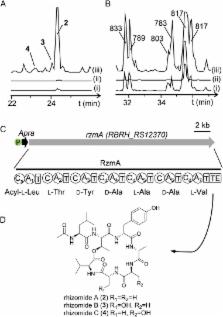
Bacterial genomes encode numerous cryptic biosynthetic gene clusters (BGCs) that represent a largely untapped source of drugs or pesticides. Mining of the cryptic products is limited by the unavailability of streamlined genetic tools in native producers. Precise genome engineering using bacteriophage recombinases is particularly useful for genome mining. However, recombinases are usually host-specific. The genome-guided discovery of novel recombinases and their transient expression could boost cryptic BGC mining. Herein, we reported a genetic system employing Red recombinases from Burkholderiales strain DSM 7029 for efficient genome engineering in several Burkholderiales species that currently lack effective genetic tools. Using specialized recombinases-assisted in situ insertion of functional promoters, we successfully mined five cryptic nonribosomal peptide synthetase/polyketide synthase BGCs, two of which were silent. Two classes of lipopeptides, glidopeptins and rhizomides, were identified through extensive spectroscopic characterization. This recombinase expression strategy offers utility within other bacteria species, allowing bioprospecting for potentially scalable discovery of novel metabolites with attractive bioactivities.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters.
- Record: found
- Abstract: found
- Article: not found
Discovery of microbial natural products by activation of silent biosynthetic gene clusters.
- Record: found
- Abstract: found
- Article: not found