- Record: found
- Abstract: found
- Article: found
Uncovering the diverse hosts of tigecycline resistance gene tet(X4) in anaerobic digestion systems treating swine manure by epicPCR

Read this article at
Highlights
-
•
A single-cell based epicPCR was developed to identify bacterial hosts of tet(X4) gene.
-
•
Nineteen genera in anaerobic digesters were identified as tet(X4) hosts by epicPCR.
-
•
Previously unknown tet(X4) hosts were found in sixteen genera.
-
•
Firmicutes and Caldatribacteriota were new phyla hosting tet(X4).
Abstract
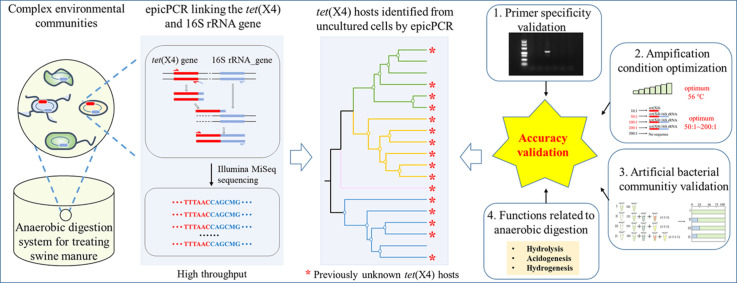
The tet(X4) gene is a clinically important tigecycline resistance gene and has shown high persistence in livestock-related environments. However, the bacterial hosts of tet(X4) remain unknown due to the lack of appropriate approaches. Herein, a culture-independent and high-throughput epicPCR (emulsion, paired isolation, and concatenation polymerase chain reaction) method was developed, optimized, and demonstrated for the identification of bacterial hosts carrying tet(X4) from environmental samples. Considering the high sequence similarity between tet(X4) and other tet(X)-variant genes, specific primers and amplification conditions were screened and optimized to identify tet(X4) accurately and link tet(X4) with the 16S rRNA gene, which were further validated using artificially constructed bacterial communities. The epicPCR targeting tet(X4) was applied for the identification of bacterial hosts carrying this resistance gene in anaerobic digestion systems treating swine manure. A total of 19 genera were identified as tet(X4) hosts, which were distributed in the phyla Proteobacteria, Bacteroidota, Firmicutes, and Caldatribacteriota. Sixteen genera and two phyla that were identified have not been previously reported as tet(X4) bacterial hosts. The results indicated that a far more diverse range of bacteria was involved in harboring tet(X4) than previously realized. Compared with the tet(X4) hosts determined by correlation-based network analysis and metagenomic binning, epicPCR revealed a high diversity of tet(X4) hosts even at the phylum level. The epicPCR method developed in this study could be effectively employed to reveal the presence of tet(X4) bacterial hosts from a holistic viewpoint.
Graphical abstract
Related collections
Most cited references74
- Record: found
- Abstract: not found
- Article: not found
Cutadapt removes adapter sequences from high-throughput sequencing reads

- Record: found
- Abstract: found
- Article: found
The SILVA ribosomal RNA gene database project: improved data processing and web-based tools
- Record: found
- Abstract: found
- Article: not found