INTRODUCTION: MALARIA PARASITES AND LIFE CYCLES
Malaria remains a serious public health burden worldwide: approximately 229 million clinical cases and 368,000 deaths were reported in 87 malaria endemic countries in 2019 [1]. Nearly 300 Plasmodium species have been characterized, infecting mammals, rodents, birds, lizards, bats, and ungulates; at least 29 species have been found to infect non-human primates [2–4] (https://en.wikipedia.org/wiki/List_of_Plasmodium_species). Human malaria is caused by five Plasmodium species: Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale, and Plasmodium knowlesi. P. falciparum is the deadliest, whereas P. vivax is the most widespread. Malaria parasites infecting a group of related vertebrate species generally have relatively strict host specificity. Plasmodium berghei, Plasmodium chabaudi, and Plasmodium yoelii, initially isolated from African thicket rats, infect only rodent hosts, such as mice and rats; in contrast, Plasmodium gallinaceum and Plasmodium relictum infect avian hosts [5,6]. The rodent or avian parasites do not infect humans and do not pose a threat to public health. However, they may have economic impacts; avian parasites infecting wild birds may be transmitted to domestic fowls and kill host animals kept in zoos or raised for food [7,8].
Many Plasmodium species can be transmitted between humans and non-human primates. For example, human infections of the New World monkey parasites Plasmodium simium and Plasmodium brasilianum have been widely reported [9]. P. simium is closely associated with P. vivax, and P. brasilianum is almost identical to P. malariae, thus suggesting transmission of these parasites between humans and New World monkeys. Recently, many potential new Plasmodium species were discovered in African apes [10–15]. Whether these parasites might infect humans, either currently or in the future, remains to be determined. Human activities expanding deeper into jungles, together with the continuing evolution of Plasmodium parasites, increases the likelihood of non-human primate parasites jumping to humans. Threats of infection of humans or domestic animals by parasites in wild animals still exist, and additional investigation on parasite evolution, adaptation, and prevention of cross-species transmission is needed.
Malaria parasites have a complex life cycle involving two hosts: female mosquitoes (Anopheles for most malaria parasites) and humans or other vertebrates as secondary hosts (Fig 1). The life cycle starts with a bite from an infected mosquito, which injects sporozoites into the skin of a vertebrate host. The sporozoites migrate within blood vesicles to the liver and infect hepatocytes [16,17]. A parasite can replicate into thousands of merozoites inside a hepatocyte, and mature merozoites are released into the bloodstream and subsequently invade erythrocytes after rupture of infected hepatocytes. Within a red blood cell (RBC), a merozoite replicates again and develops from ring to trophozoite and schizont stages. The rupture of RBCs containing schizonts also releases metabolic by-products including hemozoin from parasite digestion of hemoglobin. The parasite products trigger host immune responses and cause various clinical symptoms such as fever, chills, myalgia, headache, dizziness, and back pain [18]. For some patients, the infection may progress to severe malaria with coma (cerebral malaria), pulmonary edema, acidosis, hypoglycemia, acute renal failure, jaundice, severe anemia, and death [19–21]. Host responses, such as the release of metabolites and immune effectors, may also trigger some parasites to differentiate into male and female gametocytes [22,23]. When another mosquito consumes a bloodmeal from an infected host, the gametocytes differentiate into male and female gametes that fertilize to form zygotes in the midgut. The zygotes then develop into motile ookinetes that penetrate the mosquito midgut wall and develop into oocysts. A mature oocyst contains thousands of sporozoites that migrate to salivary glands. When a mosquito bites a vertebrate host, sporozoites are injected into the skin, and a life cycle begins. All Plasmodium species have similar life cycles with different species of mosquito and vertebrate hosts, although the life cycle length can differ. For example, the erythrocytic cycle is approximately 24 h for P. knowlesi; 48 h for P. falciparum, P. vivax, and P. ovale; and 72 h for P. malariae [19,24–27].
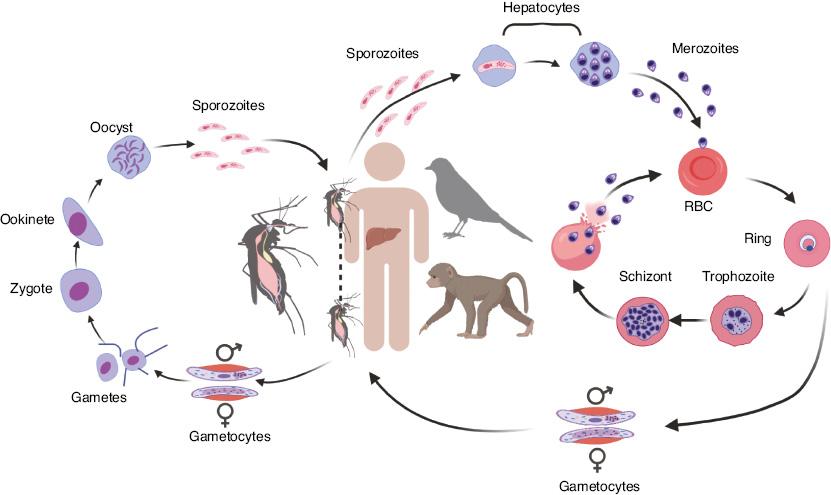
Representative life cycle of malaria parasites. Injection of sporozoites into a vertebrate host by a mosquito bite initiates the infection and the parasite life cycle. The injected sporozoites travel through the bloodstream to the liver and invade hepatocytes, where the parasites replicate and produce thousands of merozoites. Mature merozoites are again released into the bloodstream, where they invade red blood cells (RBCs). Within an RBC, the parasite replicates, producing more merozoites that can invade new RBCs for another cycle. For unknown reasons, some merozoites may develop into male and female gametocytes that differentiate into male and female gametes in the mosquito midgut when another a mosquito takes a blood meal. The male and female gametes fertilize, forming zygotes that differentiate into motile ookinetes. The ookinetes penetrate the mosquito midgut and develop into oocysts outside the midgut wall. Each oocyst contains many sporozoites that travel to the mosquito salivary glands. When the mosquito bites another vertebrate host, the sporozoites are injected into the new host, thus starting another cycle.
For this brief review, we searched PubMed and Google for selected parasite species (including P. falciparum, P. vivax, P. malariae, P. ovale, P. knowlesi, P. cynomolgi, P. simium, and P. brasilianum), together with phrases such as ‘zoonotic transmission’, ‘molecular evolution’, ‘host switch’, and ‘monkey or non-human primate malaria’. Selected relevant references cited in published articles were also reviewed and are discussed. The following section briefly describes the origins and zoonotic transmission of important malaria parasites that infect humans and non-human primates, as summarized in Fig 2. Examples of avian malaria parasites causing damage to birds in zoos and to domestic fowls are also briefly discussed.
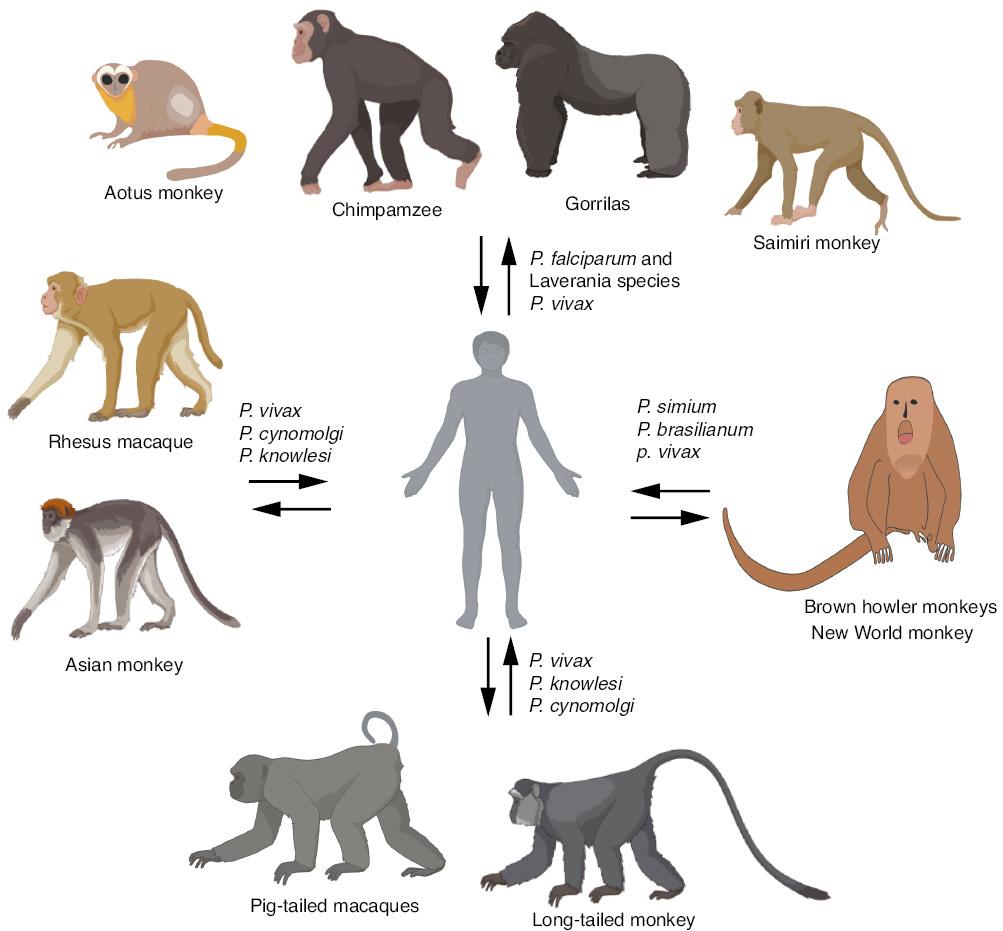
Potential transmission of malaria parasites between humans and non-human primates. Arrows indicate the direction of potential transmission of malaria parasites. The cartoons of non-human primates were created with Biorender (https://biorender.com) for illustration purposes only; they might not accurately reflect the characteristics of the species.
THE ORIGIN OF P. FALCIPARUM AND ITS TRANSMISSION BETWEEN HUMANS AND APES
With the development of DNA sequencing and genotyping tools, the origins and evolutionary histories of malaria parasites have become topics of interest, and debates are ongoing regarding the origins and times of divergence from the common ancestors of P. falciparum and P. vivax [28,29]. Analysis of small-subunit ribosomal RNA genes initially suggested that P. falciparum and avian parasites share a relatively recent progenitor [30]. The chimpanzee parasite Plasmodium reichenowi has long been considered the closest species to P. falciparum, because both parasites have high-AT content genomes and share a high degree of genome similarity [31]. According to one longstanding hypothesis, P. falciparum and P. reichenowi evolved from a P. falciparum-like common ancestor, then co-evolved with their respective hosts for millions of years [32]. Recent discoveries of a multitude of Plasmodium spp. in chimpanzees and gorillas suggest that P. falciparum probably arose from recent cross-species transmission of parasites from African apes [14,15,29,33,34]. In one study, DNA sequences in nearly 3,000 fecal samples collected from wild-living apes (chimpanzees and gorillas) were amplified and sequenced [14]. Analysis of more than 1,100 mitochondrial, apicoplast, and nuclear gene sequences from the animals grouped most of the sequences within one of six chimpanzee- or gorilla-specific lineages representing distinct Plasmodium species, including three not previously reported. Nine sequences were also associated with another group of human parasites with higher GC content (P. malariae, P. ovale, or P. vivax). These studies indicated the presence of many Plasmodium species belonging to the Laverania subgenus in the apes: P. gaboni, P. billcollinsi, and P reichenowi in chimpanzees; and P. praefalciparum, P. blackloci, and P. adleri in gorillas [14,35]. Additionally, Plasmodium lomamiensis was later identified in bonobos [15]. P. falciparum is also a member of the Laverania group and is the only parasite that has successfully adapted to humans after a potential transfer from gorillas to humans [14,35]. P. falciparum has been proposed to have arisen from a single transfer of P. praefalciparum into humans [36]. Indeed, P. falciparum is closely associated with P. praefalciparum, with an estimated divergence time of 40,000–60,000 years, and signals have indicated significant gene flow between these two parasite species after their initial divergence [35]. The jump of parasites from African apes to humans was probably due to adaption and mutations in a limited number of parasite genes involved in the invasion of RBCs and host-parasite interactions [35,37]. Parasites infecting different primate hosts appear to have various genes that are involved in either invasion or pathogenesis and have been fixed in the genomes [35]. For example, the interaction of the P. falciparum ligand EBA-175 with its erythrocyte sialoglycoprotein receptor, glycophorin A (GPA), plays a role in parasite invasion of RBCs [38]. Ape GPA sialic acid residues contain predominantly N-glycolylneuraminic acid (Neu5Gc), whereas those of humans have N-aceytlneuraminic acid (Neu5Ac). The difference in Neu5Ac and Neu5Gc determines the receptor-ligand specificity: P. falciparum EBA-175 cannot bind chimpanzee GPA, whereas the P. reichenowi homolog of EBA-175 cannot bind human GPA [39].
Although P. falciparum is known to naturally infect only humans, several splenectomized non-human primates have been shown to support P. falciparum growth and have been explored as animal models for studies of vaccine development and genetic crosses. Aotus infulatus (owl) monkeys, both intact and splenectomized, can be infected with P. falciparum [40,41]. Animals with intact spleens have low parasitemia but develop complications such as severe anemia, whereas splenectomized monkeys have substantial parasitemia without major complications. Similarly, spleen-intact and splenectomized saimiri monkeys can also be infected with the Indochina 1/CDC and Uganda Palo FUP strains of P. falciparum; however, the animals recover without developing severe disease [42]. Additionally, P. falciparum strains can be adapted to grow in splenectomized chimpanzees, which have been used as hosts for genetic crosses [43–45]. The parasites in the apes are changing and evolving, and additional parasites might potentially jump from African apes to humans, thus resulting in new human malaria parasite species in the future.
THE ORIGIN OF P. VIVAX AND ITS TRANSMISSION BETWEEN HUMANS AND NON-HUMAN PRIMATES
P. vivax is another important human malaria parasite that is endemic to Asia, Oceania, and Central/South America, but is rare in sub-Saharan Africa because of the absence of Duffy antigen on the RBCs in human populations in Africa [46]. The ongoing debate concerning the origin of P. vivax is interesting, given that data support both Asian and African origins. Several earlier studies have suggested that P. vivax might have arisen from the cross-species transmission of parasites between humans and macaques in Southeast Asia [47–50]. However, the hypothesis of Asian origin has recently been challenged after the characterization of many additional Plasmodium parasites from African apes [29]. Additional sequences from wild chimpanzees and gorillas throughout central Africa have shown that these animals were endemically infected with parasites closely associated with P. vivax, thus suggesting that P. vivax might have emerged in Africa [51]. Phylogenetic relationships derived from apicoplast genome sequences of several Asian primate parasites, Plasmodium cynomolgi, P. knowlesi, Plasmodium fragile, Plasmodium fieldi, Plasmodium simiovale, Plasmodium hylobati, Plasmodium inui, and P. gonderi also support an African origin of P. vivax [52]. However, recent analyses of genomic variation in 447 human P. vivax strains and 19 ape P. vivax-like strains collected worldwide have indicated that P. vivax might have arisen from a single area in Asia through serial founder effects [53]. P. vivax has been shown to be a sister group rather than a sublineage of P. vivax-like parasites, and a strong bottleneck in the lineage leading to P. vivax may explain the richer genetic diversity in the P. vivax-like groups [53]. Notably, before these recent analyses of large numbers of parasite isolates and genomic datasets, Richard Carter provided some potential answers to the debates on P. vivax origin, on the basis of changes in historical continental landmasses, climate, the distribution of primates over time, and malaria parasite selection on the human genome (Duffy negative) in the Old World [54,55]. Carter has speculated that P. vivax could have diverged ~2 million years ago from a group of malaria parasites that are now endemic in monkeys and apes in southern Asia, and all P. vivax today might be descended from parasites that infected human populations in the Mediterranean region and sub-Saharan Africa [55]. This hypothesis might explain the discrepant signals found in parasite sequences collected from Asian and Africa. Nonetheless, although the debate on the origin of P. vivax is expected to continue, these studies clearly illustrate the zoonotic nature of P. falciparum and P. vivax parasites.
EMERGENCE OF P. VIVAX IN AFRICA AND THE INVASION OF DUFFY NEGATIVE RBCS
An African origin of P. vivax is supported by the lack of Duffy antigen expression in African human populations and the requirement of Duffy antigen/chemokine receptor (DARC) for parasite invasion of RBCs [46,55]. The P. vivax parasite invades reticulocytes mainly through binding of P. vivax Duffy binding protein (PvDBP) to DARC [56–58]. For many years, Duffy-negative individuals were thought to be fully resistant to vivax malaria, as an example of selection of a genetic trait by an infectious disease [59]. An amino acid substitution in the GATA-1 transcription factor binding site of the gene encoding the Duffy blood group, Fy, causes an absence of DARC expression [60]. However, P. vivax has since been reported in Duffy-negative human populations [61–66]. Duplication of the PvDBP genes and low-level expression of the gene (which was previously undetectable) encoding DARC (Fy) might have contributed to the observations of P. vivax in Duffy-negative individuals [67–71]. Additionally, African great apes may act as Duffy-positive reservoirs of P. vivax in regions where P. vivax parasites (or P. vivax–like parasites) have been found in the blood of these great apes [72,73]. These observations show that new infections may occur after mutations in both host and parasite genomes, thereby leading to new transmission. However, no significant difference in PvDBP copy number has been observed in parasites infecting Duffy-null heterozygotes and Duffy-positive homozygotes/heterozygotes [74]. The molecular mechanism of infection in Duffy-negative individuals requires further investigation.
P. VIVAX/P. SIMIUM IN HUMANS AND NEW WORLD MONKEYS
P. simium can infect neotropical platyrrhine monkeys and humans. Although how P. simium specifically arrived in the Americas remains disputed, most scientists studying the parasite agree that P. simium was established in New World monkeys after migrants brought the parasite (or P. vivax initially) either from Europe and Africa during the colonial era, or from Southeast Asia [75,76]. P. vivax and P. simium have nearly identical genomes, and parasites found in humans and simians can be transmitted by the same vectors in the Atlantic Forest of Brazil [77]. More recent phylogenetic analyses based on the whole genome sequences of P. simium isolates infecting humans and brown howler monkeys support the hypothesis that P. simium first infected non-human primates as a result of a host-switch from humans carrying P. vivax [78,79]. Additionally, signals of multiple independent host switches have been detected, thus suggesting that zoonotic infections occurred in humans, and non-human primate malaria parasites serve as reservoirs for potential human infectious [79]. Indeed, an outbreak of P. simium malaria in humans in the Atlantic Forest in Rio de Janeiro has been reported [80]. Transmission from non-human primates to humans must be considered in any malaria elimination attempts in the region.
P. OVALE AND POTENTIAL ZOONOTIC TRANSMISSION
P. ovale is a human malaria parasite producing oval shaped infected erythrocytes [25,81]. Similarly to P. vivax, the parasites invade primarily reticulocytes to initiate the erythrocytic cycle, and they can suspend their development as hypnozoites in the liver for months, thus leading to relapses [25]. The parasite is found mostly in sub-Saharan Africa; islands of the western Pacific, such as the Philippines and New Guinea; the Middle East; and Southeast Asia [82–90]. Recently, two nonrecombining species of P. ovale, P. ovale curtisi (classic type) and P. ovale wallikeri (variant type), have been recognized [91,92]. Phylogenetic analysis of the Plasmodium genus on the basis of genomic sequences has placed P. ovale curtisi and P. ovale wallikeri in a clade between those of P. vivax/P. cynomolgi and P. berghei/P. chabaudi [93]. The study also supports relatively ancient divergence of the two P. ovale species, as evidenced by their different pir repertoires [93].
Genetic evidence has indicated the presence of P. ovale parasites in African chimpanzees. In a study of partial mitochondrial cytochrome b (cytb) and cytochrome c oxidase 1 (cox1) and nuclear ldh sequences from 130 samples collected from chimpanzees (Pan troglodytes) and gorillas (Gorilla gorilla) in Cameroon, P. olave sequences were detected in two chimpanzees [90]. One chimpanzee was infected with parasites whose sequences were identical to those of the variant (wallikeri) P. ovale type, and the other was infected with parasites different from the two human P. ovale parasites. In a similar study, blood samples from 12 chimpanzees and two gorillas from Cameroon and one lemur from Madagascar were examined for two parasite mitochondrial genes (cytb and cox1), one plastid gene (tufA), and ldh; P. ovale was found in the chimpanzees [94]. In another study, 16 wild West African chimpanzees in Taï National Park, Côte d’Ivoire, were found to be naturally infected with P. ovale, P. vivax, P. malariae, and three types of plasmodia rarely observed in humans in the region [95]. These observations suggest potential natural cross-species exchange in P. ovale between humans and chimpanzees.
HUMAN INFECTION OF P. MALARIAE AND P. BRASILIANUM
Simian P. brasilianum causes quartan fever in New World monkeys and resembles the human quartan parasite P. malariae under microscopic examination. P. brasilianum parasites from monkeys have been found to infect humans experimentally, and New World monkeys or chimpanzees can be experimentally infected with P. malariae from humans [96–99]. The two parasites are nearly genetically identical [9,93,100,101]. Typically, quartan malaria parasites were called P. brasilianum when identified in monkeys and P. malariae when detected in humans [101]. Analyses of parasite genes encoding the circumsporozoite protein (CSP) and the ribosomal small subunit (18S) from patients in remote Yanomami indigenous communities in Venezuela have identified 12 people harboring parasites 100% identical to P. brasilianum isolated from the monkeys, thus suggesting naturally acquired human infections of P. brasilianum [101]. Monkeys living in the rainforest therefore can act as natural reservoirs for P. brasilianum/P. malariae malaria [100]. Some studies have shown that P. brasilianum strains appear to be more divergent than P. malariae parasites [9,102], thereby suggesting that P. malariae might be derived from P. brasilianum. However, the level of genetic variation was minimal, and the diversity could represent intraspecies variation.
Genome sequence and phylogenetic analyses have shown that P. malariae and P. malariae-like make up a clade at the root of clades consisting of P. vivax/knowlesi/cynomolgi, P. ovale, and P. berghei/chabaudi [93]. Additionally, the cytb and ssrRNA sequences of P. malariae and P. malariae-like have been found in wild chimpanzees in tropical Africa [14,95], thus suggesting potential transmission of P. malariae or P. malariae-like parasites from chimpanzees to humans.
OUTBREAKS OF P. KNOWLESI INFECTIONS IN HUMAN POPULATIONS
P. knowlesi is a malaria parasite initially found in long-tailed (Macaca fascicularis) and pig-tailed (Macaca nemestrina) macaques, and human infections were thought to be rare until natural infections in many human individuals were reported in Sarawak, Malaysian Borneo, in 2004 [103,104]. The parasite was first observed in blood film from a long-tailed macaque in the early 1930s [103]. Subsequently, blood passage of the parasite in its natural host, M. fascicularis, was found to result in infection with low parasitemia [103]. Human natural infection was reported in a U.S. Army surveyor in Peninsular Malaysia more than 30 years later [105]. However, many human infections of P. knowlesi were likely to be misdiagnosed as P. malariae infections in routine microscopy, because the blood stages of P. knowlesi and P. malariae are morphologically very similar [106]. More extensive human infections of P. knowlesi have been discovered in the past 15 years, particularly with the development and application of PCR-based techniques, which have greatly improved the ability to detect P. knowlesi infection in humans and monkeys [104]. After the report of a large focus of human infections of P. knowlesi in Malaysian Borneo in 2004 [107,108], human cases of P. knowlesi infections have been reported in Malaysia [109–111] Thailand [112–114], Cambodia [115,116], Vietnam [117], Myanmar [118], the Philippines [119], Singapore [120,121], and Indonesia [122]. Importantly, the areas with natural human P. knowlesi infections overlap with the distribution of long-tailed and/or pig-tail macaques. Therefore, P. knowlesi is now recognized as the fifth human malaria species, and zoonotic transmission plays a key role in human infections. The zoonotic transmission of P. knowlesi represents a challenge for malaria control and elimination in Southeast Asia [123]. Indeed, zoonotic P. knowlesi infection has become the major cause of malaria in Malaysia, particularly Malaysian Borneo [124].
HUMAN INFECTIONS WITH THE MONKEY MALARIA PARASITE P. CYNOMOLGI
P. cynomolgi is another malaria parasite that naturally infects Asian monkey species, including long-tailed and pig-tailed macaques [125]; however, this parasite may also occasionally infect humans [126]. Human infection has been reported in two staff members who became infected with vivax-type parasites when they inoculated monkeys with P. cynomolgi sporozoites [127]. Additional transmission of P. cynomolgi from monkeys to humans or from among humans was reported shortly thereafter [128–131]. Naturally acquired human infection of P. cynomolgi was reported in a symptomatic 39-year-old female nurse in peninsular Malaysia in 2014 [132]. In a more recent survey of malaria infections involving 14,732 individuals in 23 villages in Pailin and Battambang, western Cambodia, 1,361 people were asymptomatically infected with several species of malaria parasites, including 11 people infected with P. cynomolgi, 8 people infected with P. knowlesi, and 2 people infected with both P. vivax and P. cynomolgi [116]. In Northern Sabah, Malaysia, two adult men were found to be infected with P. cynomolgi [110]. Additionally, 6 of 1,047 blood samples collected at Kapit Hospital in Kapit, Malaysia, were found to be co-infected with P. cynomolgi and P. knowlesi [111]. These reports suggest that P. cynomolgi can also infect humans naturally, although the prevalence of P. cynomolgi infection in humans may be lower than that of P. knowlesi. More outbreaks of P. cynomolgi infection might potentially be undetected or could occur in the future if the parasites acquire mutations in the key parasite genes involved in binding to host receptors.
MALARIA SPECIES IN BIRDS AND THEIR TRANSMISSION BETWEEN CAPTIVE/DOMESTIC AND WILD BIRDS
Avian hosts are infected with many species of malaria parasites such as P. relictum, Plasmodium elongatum, Plasmodium juxtanucleare, and P. gallinaceum [6,133]. Although these parasites do not infect humans and are generally not of concern to public health, transmission of parasites from wild birds to domestic fowls can inflict economic losses on the poultry industry. Transmission of many avian malaria species is mediated by species of Culicidae mosquitoes in several genera (Culex, Coquillettidia, Aedes, Mansonia, Culisetta, Anopheles, and Psorophora), instead of the Anopheles species for mammalian malaria parasites [134,135]. Many species of avian malaria parasites exist among wild birds, but studies on the disease severity and pathology in wild birds are limited [136,137]. Traditionally, wild birds infected with these parasites were generally considered to have mild disease. However, avian malaria has been shown to play a role in the ongoing decline of native New Zealand birds [138]. Diminished overwinter survival of both juvenile and adult house sparrows has been associated with the intensity of Plasmodium infections [139]. Various clinical signs have been observed in domestic chickens after infection with P. gallinaceum, including depression, fever, anorexia, reduced weight gain, poor feed conversion, anemia, green feces, and even death [140]. Death of experimental passerine birds due to marked organ damage has been reported after Plasmodium homocircumflexum infection [137]. Avian malaria, most commonly associated with P. relictum or P. elongatum, can also cause severe disease in penguins, with mortality rates as high as 50%–80% [141]. Vector and bird migration and vector introduction by human action into non-endemic habitats represents a risk of endangering species [142]. For example, P. relictum has recently been reported to expand its range, infecting two non-migratory passerines in North America [143]. Species of Plasmodium, Haemoproteus, and Leucocytozoon have been reported to infect cranes in the Beijing Zoo, particularly the juvenile birds that often die from infection by Plasmodium and Leucocytozoon parasites [144]. Therefore, the transmission of avian malaria parasites between wild birds and domestic or captive avian hosts can be considered examples of zoonotic transmission of malaria parasites.
CONCLUSION
The transmission of malaria parasites from non-human primates to humans and vice versa has occurred many times during the evolutionary histories of malaria parasites and their hosts. Moreover, some parasites have adapted to develop in both humans and non-human primates. All five malaria parasite species that infect humans have been found in non-human primates, which in turn can act as reservoirs for transmission to humans. Additionally, the malaria species currently infecting wild non-human primates may gain mutations that allow them to bind receptors in humans and subsequently jump to humans. Furthermore, many unknown species in wildlife hosts remain to be characterized. One approach might be to collect fecal or tissue (blood) samples from potential hosts such as bats, wild rodents, and non-human primates, and extract DNA/RNA for high throughput sequencing to reveal parasite genetic information for species characterization.
The risk of malaria parasite outbreaks is a frequent threat to public health when human activities lead to close contact with non-human primates. Close surveillance and more studies on the biology and transmission of parasites circulating among non-human primates are of critical importance for preventing outbreaks of malaria due to zoonotic infection.