- Record: found
- Abstract: found
- Article: found
Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing

Read this article at
Abstract
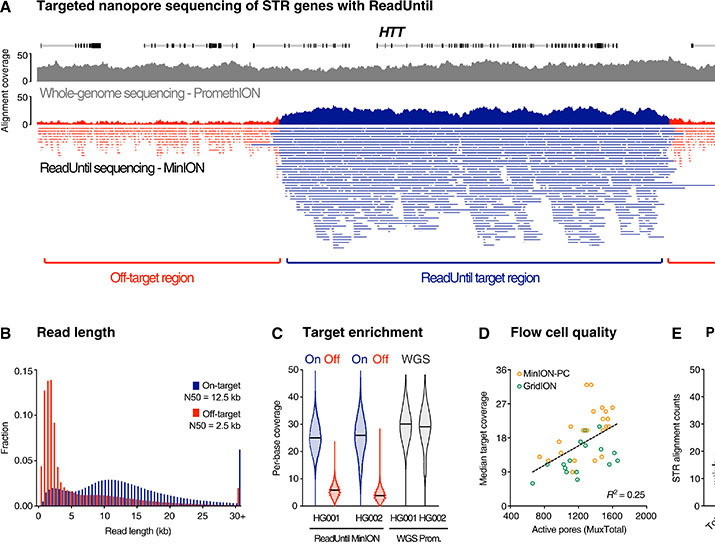
More than 50 neurological and neuromuscular diseases are caused by short tandem repeat (STR) expansions, with 37 different genes implicated to date. We describe the use of programmable targeted long-read sequencing with Oxford Nanopore’s ReadUntil function for parallel genotyping of all known neuropathogenic STRs in a single assay. Our approach enables accurate, haplotype-resolved assembly and DNA methylation profiling of STR sites, from a list of predetermined candidates. This correctly diagnoses all individuals in a small cohort ( n = 37) including patients with various neurogenetic diseases ( n = 25). Targeted long-read sequencing solves large and complex STR expansions that confound established molecular tests and short-read sequencing and identifies noncanonical STR motif conformations and internal sequence interruptions. We observe a diversity of STR alleles of known and unknown pathogenicity, suggesting that long-read sequencing will redefine the genetic landscape of repeat disorders. Last, we show how the inclusion of pharmacogenomic genes as secondary ReadUntil targets can further inform patient care.
Abstract
Abstract
Programmable targeted nanopore sequencing profiles all known pathogenic short tandem repeats (STRs) in a single test.
Related collections
Most cited references87
- Record: found
- Abstract: found
- Article: not found
Minimap2: pairwise alignment for nucleotide sequences
- Record: found
- Abstract: found
- Article: not found
Assembly of long, error-prone reads using repeat graphs
- Record: found
- Abstract: found
- Article: not found