- Record: found
- Abstract: found
- Article: not found
Polyelectrolyte in Electric Field: Disparate Conformational Behavior along an Aminopolysaccharide Chain
Read this article at
Abstract

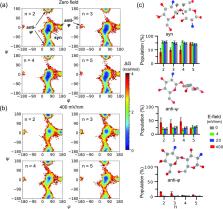
Electrical signals are increasingly used in fabrication of hydrogels (e.g., based on aminopolysaccharide chitosan) to guide the emergence of complex and anisotropic structure; however, how an imposed electric field affects the polymer chain conformation and orientation during the self-assembly process is not understood. Here, we applied nonequilibrium all-atom molecular dynamics simulations to explore the response of a charged chitosan chain comprising 5- or 20-monomer units to a constant uniform electric field in water and salt solution. While no conformational or orientational response was observed for the polyelectrolyte (PE) chains under the small electric fields within the simulation time, a field strength of 400 mV/nm induced significant changes. In water, a 5-mer chain is found to be slightly bent and oriented parallel to the field; however, surprisingly, a 20-mer chain displays candy-cane-like conformations whereby one half of the chain is collapsed and flexible, while the other half of the chain is stretched along the electric field. In salt solution, the disparity remains between the two halves of the 20-mer chain, although the backbone is extremely flexible with multiple bent regions and non-native conformations occur near the chain center in one of the three trajectories. The disparate conformational response along the polyelectrolyte chain may be attributed to the balancing forces between chain dynamics, electric polarization, counterion binding, and hydrodynamic pressure as well as friction. These findings reconcile existing experiments and theoretical studies and represent an important step toward understanding the complex roles of electric field and salt in controlling the structure and properties of soft matter.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling.
- Record: found
- Abstract: found
- Article: not found
Biofabrication with chitosan.
- Record: found
- Abstract: found
- Article: not found
