- Record: found
- Abstract: found
- Article: found
Comparison of the pathogenesis of SARS-CoV-2 infection in K18-hACE2 mouse and Syrian golden hamster models

Read this article at
ABSTRACT
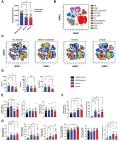
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of COVID-19, causes life-threatening disease. This novel coronavirus enters host cells via the respiratory tract, promoting the formation of severe pulmonary lesions and systemic disease. Few animal models can simulate the clinical signs and pathology of COVID-19 patients. Diverse preclinical studies using K18-hACE2 mice and Syrian golden hamsters, which are highly permissive to SARS-CoV-2 in the respiratory tract, are emerging; however, the systemic pathogenesis and cellular tropism of these models remain obscure. We intranasally infected K18-hACE2 mice and Syrian golden hamsters with SARS-CoV-2, and compared the clinical features, pathogenesis, cellular tropism and infiltrated immune-cell subsets. In K18-hACE2 mice, SARS-CoV-2 persistently replicated in alveolar cells and caused pulmonary and extrapulmonary disease, resulting in fatal outcomes. Conversely, in Syrian golden hamsters, transient SARS-CoV-2 infection in bronchial cells caused reversible pulmonary disease, without mortality. Our findings provide comprehensive insights into the pathogenic spectrum of COVID-19 using preclinical models.
Abstract
[Related article:] Editor's choice: We infected K18-hACE2 mice and Syrian golden hamsters with SARS-CoV-2 and obtained comprehensive insights into COVID-19 pathogenicity in these two representative COVID-19 preclinical models.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study
- Record: found
- Abstract: found
- Article: not found
Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19
- Record: found
- Abstract: found
- Article: found