- Record: found
- Abstract: found
- Article: not found
Porcine Coronaviruses: Overview of the State of the Art
Read this article at
Abstract
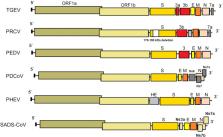
Like RNA viruses in general, coronaviruses (CoV) exhibit high mutation rates which, in combination with their strong tendency to recombine, enable them to overcome the host species barrier and adapt to new hosts. It is currently known that six CoV are able to infect pigs. Four of them belong to the genus Alphacoronavirus [transmissible gastroenteritis coronavirus (TEGV), porcine respiratory coronavirus (PRCV), porcine epidemic diarrhea virus (PEDV), swine acute diarrhea syndrome coronavirus (SADS-CoV)], one of them to the genus Betacoronavirus [porcine hemagglutinating encephalomyelitis virus (PHEV)] and the last one to the genus Deltacoronavirus (PDCoV). PHEV was one of the first identified swine CoV and is still widespread, causing subclinical infections in pigs in several countries. PRCV, a spike deletion mutant of TGEV associated with respiratory tract infection, appeared in the 1980s. PRCV is considered non-pathogenic since its infection course is mild or subclinical. Since its appearance, pig populations have become immune to both PRCV and TGEV, leading to a significant reduction in the clinical and economic importance of TGEV. TGEV, PEDV and PDCoV are enteropathogenic CoV and cause clinically indistinguishable acute gastroenteritis in all age groups of pigs. PDCoV and SADS-CoV have emerged in 2014 (US) and in 2017 (China), respectively. Rapid diagnosis is crucial for controlling CoV infections and preventing them from spreading. Since vaccines are available only for some porcine CoV, prevention should focus mainly on a high level of biosecurity . In view of the diversity of CoV and the potential risk factors associated with zoonotic emergence, updating the knowledge concerning this area is essential.
Related collections
Most cited references150
- Record: found
- Abstract: found
- Article: found
Emerging coronaviruses: Genome structure, replication, and pathogenesis
- Record: found
- Abstract: found
- Article: not found
Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus.
- Record: found
- Abstract: found
- Article: not found
