- Record: found
- Abstract: found
- Article: found
Staphylococcus aureus Entrance into the Dairy Chain: Tracking S. aureus from Dairy Cow to Cheese
Read this article at
Abstract
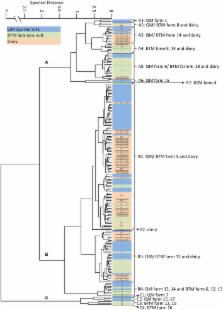
Staphylococcus aureus is one of the most important contagious mastitis pathogens in dairy cattle. Due to its zoonotic potential, control of S. aureus is not only of great economic importance in the dairy industry but also a significant public health concern. The aim of this study was to decipher the potential of bovine udder associated S. aureus as reservoir for S. aureus contamination in dairy production and processing. From 18 farms, delivering their milk to an alpine dairy plant for the production of smeared semi-hard and hard cheese. one thousand hundred seventy six one thousand hundred seventy six quarter milk (QM) samples of all cows in lactation ( n = 294) and representative samples form bulk tank milk (BTM) of all farms were surveyed for coagulase positive (CPS) and coagulase negative Staphylococci (CNS). Furthermore, samples from different steps of the cheese manufacturing process were tested for CPS and CNS. As revealed by chemometric-assisted FTIR spectroscopy and molecular subtyping ( spa typing and multi locus sequence typing), dairy cattle represent indeed an important, yet underreported, entrance point of S. aureus into the dairy chain. Our data clearly show that certain S. aureus subtypes are present in primary production as well as in the cheese processing at the dairy plant. However, although a considerable diversity of S. aureus subtypes was observed in QM and BTM at the farms, only certain S. aureus subtypes were able to enter and persist in the cheese manufacturing at the dairy plant and could be isolated from cheese until day 14 of ripening. Farm strains belonging to the FTIR cluster B1 and B3, which show genetic characteristics (t2953, ST8, enterotoxin profile: sea/ sed/ sej) of the recently described S. aureus genotype B, most successfully contaminated the cheese production at the dairy plant. Thus, our study fosters the hypothesis that genotype B S. aureus represent a specific challenge in control of S. aureus in the dairy chain that requires effective clearance strategies and hygienic measures already in primary production to avoid a potential transfer of enterotoxic strains or enterotoxins into the dairy processing and the final retail product.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
Staphylococcal enterotoxins.
- Record: found
- Abstract: found
- Article: not found
Microbiological characterizations by FT-IR spectroscopy.
- Record: found
- Abstract: found
- Article: not found