- Record: found
- Abstract: found
- Article: found
17q12 deletion syndrome mouse model shows defects in craniofacial, brain and kidney development, and glucose homeostasis
Read this article at
ABSTRACT
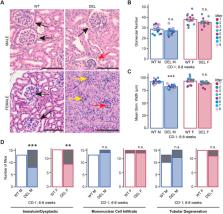
17q12 deletion (17q12Del) syndrome is a copy number variant (CNV) disorder associated with neurodevelopmental disorders and renal cysts and diabetes syndrome (RCAD). Using CRISPR/Cas9 genome editing, we generated a mouse model of 17q12Del syndrome on both inbred (C57BL/6N) and outbred (CD-1) genetic backgrounds. On C57BL/6N, the 17q12Del mice had severe head development defects, potentially mediated by haploinsufficiency of Lhx1, a gene within the interval that controls head development. Phenotypes included brain malformations, particularly disruption of the telencephalon and craniofacial defects. On the CD-1 background, the 17q12Del mice survived to adulthood and showed milder craniofacial and brain abnormalities. We report postnatal brain defects using automated magnetic resonance imaging-based morphometry. In addition, we demonstrate renal and blood glucose abnormalities relevant to RCAD. On both genetic backgrounds, we found sex-specific presentations, with male 17q12Del mice exhibiting higher penetrance and more severe phenotypes. Results from these experiments pinpoint specific developmental defects and pathways that guide clinical studies and a mechanistic understanding of the human 17q12Del syndrome. This mouse mutant represents the first and only experimental model to date for the 17q12 CNV disorder.
This article has an associated First Person interview with the first author of the paper.
Abstract
Summary: We developed a mouse model of 17q12 deletion syndrome, which phenocopies many of the features of the human disorder, including renal dysfunction, craniofacial, glucose homeostasis and brain abnormalities.
Related collections
Most cited references118
- Record: found
- Abstract: not found
- Article: not found
Fitting Linear Mixed-Effects Models Usinglme4
- Record: found
- Abstract: not found
- Article: not found
lmerTest Package: Tests in Linear Mixed Effects Models
- Record: found
- Abstract: found
- Article: not found