- Record: found
- Abstract: found
- Article: found
Multiscale patterns and drivers of arbuscular mycorrhizal fungal communities in the roots and root‐associated soil of a wild perennial herb
Read this article at
Summary
-
Arbuscular mycorrhizal (AM) fungi form diverse communities and are known to influence above‐ground community dynamics and biodiversity. However, the multiscale patterns and drivers of AM fungal composition and diversity are still poorly understood.
-
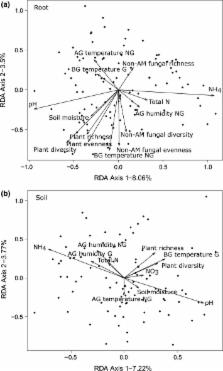
We sequenced DNA markers from roots and root‐associated soil from Plantago lanceolata plants collected across multiple spatial scales to allow comparison of AM fungal communities among neighbouring plants, plant subpopulations, nearby plant populations, and regions. We also measured soil nutrients, temperature, humidity, and community composition of neighbouring plants and nonAM root‐associated fungi.
-
AM fungal communities were already highly dissimilar among neighbouring plants ( c. 30 cm apart), albeit with a high variation in the degree of similarity at this small spatial scale. AM fungal communities were increasingly, and more consistently, dissimilar at larger spatial scales. Spatial structure and environmental drivers explained a similar percentage of the variation, from 7% to 25%. A large fraction of the variation remained unexplained, which may be a result of unmeasured environmental variables, species interactions and stochastic processes.
-
We conclude that AM fungal communities are highly variable among nearby plants. AM fungi may therefore play a major role in maintaining small‐scale variation in community dynamics and biodiversity.
Abstract
See also the Commentary on this article by https://doi.org/10.1111/nph.15212.
Related collections
Most cited references61
- Record: found
- Abstract: not found
- Book Chapter: not found
AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS
- Record: found
- Abstract: found
- Article: not found
Patterns and processes of microbial community assembly.
- Record: found
- Abstract: found
- Article: not found