- Record: found
- Abstract: found
- Article: found
Bon-EV: an improved multiple testing procedure for controlling false discovery rates
Read this article at
Abstract
Background
Stability of multiple testing procedures, defined as the standard deviation of total number of discoveries, can be used as an indicator of variability of multiple testing procedures. Improving stability of multiple testing procedures can help to increase the consistency of findings from replicated experiments. Benjamini-Hochberg’s and Storey’s q-value procedures are two commonly used multiple testing procedures for controlling false discoveries in genomic studies. Storey’s q-value procedure has higher power and lower stability than Benjamini-Hochberg’s procedure. To improve upon the stability of Storey’s q-value procedure and maintain its high power in genomic data analysis, we propose a new multiple testing procedure, named Bon-EV, to control false discovery rate (FDR) based on Bonferroni’s approach.
Results
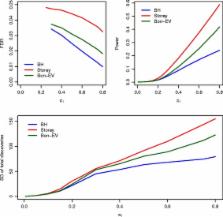
Simulation studies show that our proposed Bon-EV procedure can maintain the high power of the Storey’s q-value procedure and also result in better FDR control and higher stability than Storey’s q-value procedure for samples of large size(30 in each group) and medium size (15 in each group) for either independent, somewhat correlated, or highly correlated test statistics. When sample size is small (5 in each group), our proposed Bon-EV procedure has performance between the Benjamini-Hochberg procedure and the Storey’s q-value procedure. Examples using RNA-Seq data show that the Bon-EV procedure has higher stability than the Storey’s q-value procedure while maintaining equivalent power, and higher power than the Benjamini-Hochberg’s procedure.
Conclusions
For medium or large sample sizes, the Bon-EV procedure has improved FDR control and stability compared with the Storey’s q-value procedure and improved power compared with the Benjamini-Hochberg procedure. The Bon-EV multiple testing procedure is available as the BonEV package in R for download at https://CRAN.R-project.org/package=BonEV.
Related collections
Most cited references11
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: not found
- Article: not found
On the Adaptive Control of the False Discovery Rate in Multiple Testing With Independent Statistics
- Record: found
- Abstract: not found
- Article: not found