- Record: found
- Abstract: found
- Article: found
Arms race between anti‐silencing and RdDM in noncoding regions of transposable elements
Read this article at
Abstract
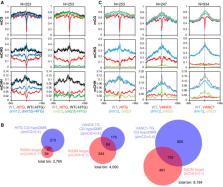
Transposable elements (TEs) are among the most dynamic parts of genomes. Since TEs are potentially deleterious, eukaryotes silence them through epigenetic mechanisms such as repressive histone modifications and DNA methylation. We previously reported that Arabidopsis TEs, called VANDALs, counteract epigenetic silencing through a group of sequence‐specific anti‐silencing proteins, VANCs. VANC proteins bind to noncoding regions of specific VANDAL copies and induce loss of silent chromatin marks. The VANC‐target regions form tandem repeats, which diverge rapidly. Sequence‐specific anti‐silencing allows these TEs to proliferate with minimum host damage. Here, we show that RNA‐directed DNA methylation (RdDM) efficiently targets noncoding regions of VANDAL TEs to silence them de novo. Thus, escape from RdDM could be a primary event leading to the rapid evolution and diversification of sequence‐specific anti‐silencing systems. We propose that this selfish behavior of TEs paradoxically could make them diverse and less harmful to the host.
Abstract
Related collections
Most cited references68

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools

- Record: found
- Abstract: found
- Article: found
