- Record: found
- Abstract: found
- Article: found
Heterologous prime-boost vaccination drives early maturation of HIV broadly neutralizing antibody precursors in humanized mice
Read this article at
Abstract
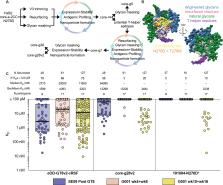
A protective human immunodeficiency virus (HIV) vaccine will likely need to induce broadly neutralizing antibodies (bnAbs). Vaccination with the germline-targeting immunogen eOD-GT8 60mer adjuvanted with AS01 B was found to induce VRC01-class bnAb precursors in 97% of vaccine recipients in the IAVI G001 phase 1 clinical trial; however, heterologous boost immunizations with antigens more similar to the native glycoprotein will be required to induce bnAbs. Therefore, we designed core-g28v2 60mer, a nanoparticle immunogen to be used as a first boost following eOD-GT8 60mer priming. We found, using a humanized mouse model approximating human conditions of VRC01-class precursor B cell diversity, affinity, and frequency, that both protein- and mRNA-based heterologous prime-boost regimens induced VRC01-class antibodies that gained key mutations and bound to near-native HIV envelope trimers lacking the N276 glycan. We further showed that VRC01-class antibodies induced by mRNA-based regimens could neutralize pseudoviruses lacking the N276 glycan. These results demonstrated that heterologous boosting can drive maturation toward VRC01-class bnAb development and supported the initiation of the IAVI G002 phase 1 trial testing mRNA-encoded nanoparticle prime-boost regimens.
One Sentence Summary:
An HIV vaccine first-boost candidate immunogen promoted maturation of VRC01-class antibodies in a humanized mouse model, supporting clinical testing.
Editor’s Summary:
Boosting HIV Antibodies. For a vaccine regimen to be effective against human immunodeficiency virus (HIV) infection, it will need to elicit broadly neutralizing antibodies (bnAbs). However, strategies to elicit bnAbs in humans with a vaccine have not been successful to date. A different strategy, germline-targeting vaccine design, relies on a priming immunogen to first induce bnAb precursors and then a series of heterologous boosters to drive or shepherd the maturation of bnAb precursors to produce bnAbs. The first step in this process, elicitation of bnAb precursors, has proven successful in clinical trials. Here, Cottrell et al. take the next step by designing and testing the first booster immunogen in the shepherding process. Vaccination of eOD-GT8 60mer-primed humanized mice with their immunogen, core-g28v2 60mer, in either protein or mRNA form resulted in antibodies that were closer to bnAbs than those that received a placebo booster. These data support the ongoing clinical trial testing core-g28v2 60mer mRNA as a booster vaccine. –Courtney Malo
Related collections
Most cited references54
- Record: found
- Abstract: found
- Article: not found
Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine
- Record: found
- Abstract: found
- Article: not found
Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine
- Record: found
- Abstract: found
- Article: not found