- Record: found
- Abstract: found
- Article: found
Landscape genomics reveals genetic signals of environmental adaptation of African wild eggplants

Read this article at
Abstract
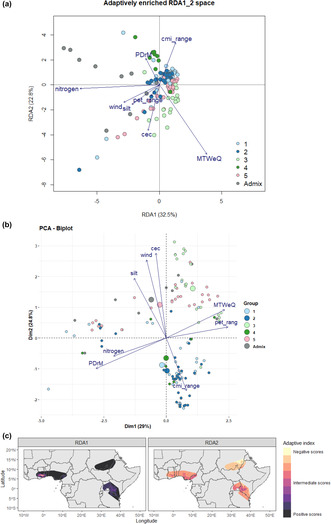
Crop wild relatives (CWR) provide a valuable resource for improving crops. They possess desirable traits that confer resilience to various environmental stresses. To fully utilize crop wild relatives in breeding and conservation programs, it is important to understand the genetic basis of their adaptation. Landscape genomics associates environments with genomic variation and allows for examining the genetic basis of adaptation. Our study examined the differences in allele frequency of 15,416 single nucleotide polymorphisms (SNPs) generated through genotyping by sequencing approach among 153 accessions of 15 wild eggplant relatives and two cultivated species from Africa, the principal hotspot of these wild relatives. We also explored the correlation between these variations and the bioclimatic and soil conditions at their collection sites, providing a comprehensive understanding of the genetic signals of environmental adaptation in African wild eggplant. Redundancy analysis (RDA) results showed that the environmental variation explained 6% while the geographical distances among the collection sites explained 15% of the genomic variation in the eggplant wild relative populations when controlling for population structure. Our findings indicate that even though environmental factors are not the main driver of selection in eggplant wild relatives, it is influential in shaping the genomic variation over time. The selected environmental variables and candidate SNPs effectively revealed grouping patterns according to the environmental characteristics of sampling sites. Using four genotype–environment association methods, we detected 396 candidate SNPs (2.5% of the initial SNPs) associated with eight environmental factors. Some of these SNPs signal genes involved in pathways that help adapt to environmental stresses such as drought, heat, cold, salinity, pests, and diseases. These candidate SNPs will be useful for marker‐assisted improvement and characterizing the germplasm of this crop for developing climate‐resilient eggplant varieties. The study provides a model for applying landscape genomics to other crops' wild relatives.
Abstract
Related collections
Most cited references115

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools

- Record: found
- Abstract: found
- Article: found
Fast and accurate short read alignment with Burrows–Wheeler transform
- Record: found
- Abstract: not found
- Article: not found