- Record: found
- Abstract: found
- Article: found
Bacterial Manipulation of Wnt Signaling: A Host-Pathogen Tug-of-Wnt
Read this article at
Abstract
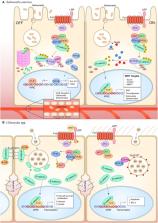
The host-pathogen interface is a crucial battleground during bacterial infection in which host defenses are met with an array of bacterial counter-mechanisms whereby the invader aims to make the host environment more favorable to survival and dissemination. Interestingly, the eukaryotic Wnt signaling pathway has emerged as a key player in the host and pathogen tug-of-war. Although studied for decades as a regulator of embryogenesis, stem cell maintenance, bone formation, and organogenesis, Wnt signaling has recently been shown to control processes related to bacterial infection in the human host. Wnt signaling pathways contribute to cell cycle control, cytoskeleton reorganization during phagocytosis and cell migration, autophagy, apoptosis, and a number of inflammation-related events. Unsurprisingly, bacterial pathogens have evolved strategies to manipulate these Wnt-associated processes in order to enhance infection and survival within the human host. In this review, we examine the different ways human bacterial pathogens with distinct host cell tropisms and lifestyles exploit Wnt signaling for infection and address the potential of harnessing Wnt-related mechanisms to combat infectious disease.
Related collections
Most cited references134
- Record: found
- Abstract: found
- Article: not found
Autophagy defends cells against invading group A Streptococcus.
- Record: found
- Abstract: found
- Article: not found
Dishevelled: The hub of Wnt signaling.
- Record: found
- Abstract: not found
- Article: not found