- Record: found
- Abstract: found
- Article: found
Functional Variants in NFKBIE and RTKN2 Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
Read this article at
Abstract
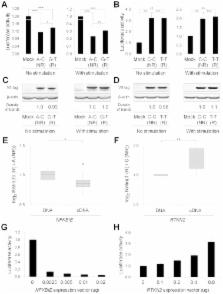
Rheumatoid arthritis is an autoimmune disease with a complex etiology, leading to inflammation of synovial tissue and joint destruction. Through a genome-wide association study (GWAS) and two replication studies in the Japanese population (7,907 cases and 35,362 controls), we identified two gene loci associated with rheumatoid arthritis susceptibility ( NFKBIE at 6p21.1, rs2233434, odds ratio (OR) = 1.20, P = 1.3×10 −15; RTKN2 at 10q21.2, rs3125734, OR = 1.20, P = 4.6×10 −9). In addition to two functional non-synonymous SNPs in NFKBIE, we identified candidate causal SNPs with regulatory potential in NFKBIE and RTKN2 gene regions by integrating in silico analysis using public genome databases and subsequent in vitro analysis. Both of these genes are known to regulate the NF-κB pathway, and the risk alleles of the genes were implicated in the enhancement of NF-κB activity in our analyses. These results suggest that the NF-κB pathway plays a role in pathogenesis and would be a rational target for treatment of rheumatoid arthritis.
Author Summary
Rheumatoid arthritis (RA) is a chronic autoimmune disease affecting approximately 1% of the general adult population. More than 30 susceptibility loci for RA have been identified through genome-wide association studies (GWAS), but the disease-causal variants at most loci remain unknown. Here, we performed replication studies of the candidate loci of our previous GWAS using Japanese cohorts and identified variants in NFKBIE and RTKN2 gene loci that were associated with RA. To search for causal variants in both gene regions, we first examined non-synonymous (ns)SNPs that alter amino-acid sequences. As NFKBIE and RTKN2 are known to be involved in the NF-κB pathway, we evaluated the effects of nsSNPs on NF-κB activity. Next, we screened in silico variants that may regulate gene transcription using publicly available epigenetic databases and subsequently evaluated their regulatory potential using in vitro assays. As a result, we identified multiple candidate causal variants in NFKBIE (2 nsSNPs and 1 regulatory SNP) and RTKN2 (2 regulatory SNPs), indicating that our integrated in silico and in vitro approach is useful for the identification of causal variants in the post–GWAS era.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis.
- Record: found
- Abstract: found
- Article: not found
Genotype imputation.
- Record: found
- Abstract: found
- Article: not found