- Record: found
- Abstract: found
- Article: found
Surface plasmon resonance immunosensor for label-free detection of BIRC5 biomarker in spontaneously occurring canine mammary tumours

Read this article at
Abstract
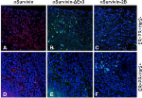
We report detection of Baculoviral inhibitor of apoptosis repeat containing-5 (BIRC5) protein biomarker in dog serum by label-free surface plasmon resonance (SPR) immunosensor. Initially, overexpression of BIRC5 in canine mammary tumour (CMT) tissues was confirmed by real-time PCR. Recombinant BIRC5 was produced and protein specific antibodies developed in guinea pig specifically reacted with native protein in immunohistochemistry and immunocytochemistry. SPR immunosensor was developed by fabricating anti-BIRC5 antibodies on gold sensor disc. The equilibrium dissociation constant, (K D = k d/k a) was 12.1 × 10 −12 M; which indicates that antibodies are of high affinity with sensitivity in picomolar range. The SPR assay could detect as low as 6.25 pg/ml of BIRC5 protein in a calibration experiment (r 2 = 0.9964). On testing real clinical samples, 95% specificity and 73.33% sensitivity were recorded. The average amount of serum BIRC5 in dogs with CMT was 110.02 ± 9.77 pg/ml; whereas, in non-cancerous disease conditions, 44.79 ± 4.28 pg/ml and in healthy dog sera 30.28 ± 2.99 pg/ml protein was detected. The SPR immunosensor for detection of BIRC5 in dog sera is reported for the first time and this may find prognostic and diagnostic applications in management of CMT. In future, ‘on-site’ sensors can be developed using this technique for near-patient testing.
Related collections
Most cited references52
- Record: found
- Abstract: not found
- Article: not found
Surface plasmon resonance sensors for detection of chemical and biological species.
- Record: found
- Abstract: found
- Article: not found
Progress and challenges in screening for early detection of ovarian cancer.
- Record: found
- Abstract: found
- Article: found