- Record: found
- Abstract: found
- Article: found
Dynamics of microbial populations mediating biogeochemical cycling in a freshwater lake
Read this article at
Abstract
Background
Microbial processes are intricately linked to the depletion of oxygen in in-land and coastal water bodies, with devastating economic and ecological consequences. Microorganisms deplete oxygen during biomass decomposition, degrading the habitat of many economically important aquatic animals. Microbes then turn to alternative electron acceptors, which alter nutrient cycling and generate potent greenhouse gases. As oxygen depletion is expected to worsen with altered land use and climate change, understanding how chemical and microbial dynamics impact dead zones will aid modeling efforts to guide remediation strategies. More work is needed to understand the complex interplay between microbial genes, populations, and biogeochemistry during oxygen depletion.
Results
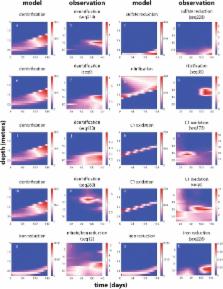
Here, we used 16S rRNA gene surveys, shotgun metagenomic sequencing, and a previously developed biogeochemical model to identify genes and microbial populations implicated in major biogeochemical transformations in a model lake ecosystem. Shotgun metagenomic sequencing was done for one time point in Aug., 2013, and 16S rRNA gene sequencing was done for a 5-month time series (Mar.–Aug., 2013) to capture the spatiotemporal dynamics of genes and microorganisms mediating the modeled processes. Metagenomic binning analysis resulted in many metagenome-assembled genomes (MAGs) that are implicated in the modeled processes through gene content similarity to cultured organism and the presence of key genes involved in these pathways. The MAGs suggested some populations are capable of methane and sulfide oxidation coupled to nitrate reduction. Using the model, we observe that modulating these processes has a substantial impact on overall lake biogeochemistry. Additionally, 16S rRNA gene sequences from the metagenomic and amplicon libraries were linked to processes through the MAGs. We compared the dynamics of microbial populations in the water column to the model predictions. Many microbial populations involved in primary carbon oxidation had dynamics similar to the model, while those associated with secondary oxidation processes deviated substantially.
Conclusions
This work demonstrates that the unique capabilities of resident microbial populations will substantially impact the concentration and speciation of chemicals in the water column, unless other microbial processes adjust to compensate for these differences. It further highlights the importance of the biological aspects of biogeochemical processes, such as fluctuations in microbial population dynamics. Integrating gene and population dynamics into biogeochemical models has the potential to improve predictions of the community response under altered scenarios to guide remediation efforts.
Related collections
Most cited references53
- Record: found
- Abstract: not found
- Article: not found
Ferrozine---a new spectrophotometric reagent for iron
- Record: found
- Abstract: found
- Article: not found
Patterns and processes of microbial community assembly.
- Record: found
- Abstract: not found
- Article: not found