- Record: found
- Abstract: found
- Article: found
CRISPR/Cas9-gene editing approaches in plant breeding
Read this article at
ABSTRACT
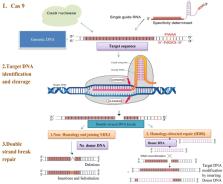
CRISPR/Cas9 gene editing system is recently developed robust genome editing technology for accelerating plant breeding. Various modifications of this editing system have been established for adaptability in plant varieties as well as for its improved efficiency and portability. This review provides an in-depth look at the various strategies for synthesizing gRNAs for efficient delivery in plant cells, including chemical synthesis and in vitro transcription. It also covers traditional analytical tools and emerging developments in detection methods to analyze CRISPR/Cas9 mediated mutation in plant breeding. Additionally, the review outlines the various analytical tools which are used to detect and analyze CRISPR/Cas9 mediated mutations, such as next-generation sequencing, restriction enzyme analysis, and southern blotting. Finally, the review discusses emerging detection methods, including digital PCR and qPCR. Hence, CRISPR/Cas9 has great potential for transforming agriculture and opening avenues for new advancements in the system for gene editing in plants.
Related collections
Most cited references149
- Record: found
- Abstract: found
- Article: not found
A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.
- Record: found
- Abstract: found
- Article: not found
Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9
- Record: found
- Abstract: found
- Article: not found