- Record: found
- Abstract: found
- Article: found
Review of Single-Cell RNA Sequencing in the Heart
Read this article at
Abstract
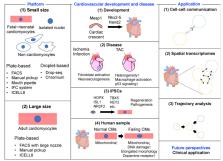
Single-cell RNA sequencing (scRNA-seq) technology is a powerful, rapidly developing tool for characterizing individual cells and elucidating biological mechanisms at the cellular level. Cardiovascular disease is one of the major causes of death worldwide and its precise pathology remains unclear. scRNA-seq has provided many novel insights into both healthy and pathological hearts. In this review, we summarize the various scRNA-seq platforms and describe the molecular mechanisms of cardiovascular development and disease revealed by scRNA-seq analysis. We then describe the latest technological advances in scRNA-seq. Finally, we discuss how to translate basic research into clinical medicine using scRNA-seq technology.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets.
- Record: found
- Abstract: found
- Article: not found
RNA velocity of single cells
- Record: found
- Abstract: found
- Article: not found