- Record: found
- Abstract: found
- Article: found
Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis
Read this article at
Abstract
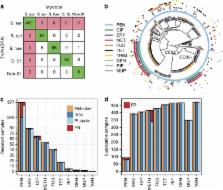
The rise of antibiotic-resistant bacteria has led to an urgent need for rapid detection of drug resistance in clinical samples, and improvements in global surveillance. Here we show how de Bruijn graph representation of bacterial diversity can be used to identify species and resistance profiles of clinical isolates. We implement this method for Staphylococcus aureus and Mycobacterium tuberculosis in a software package (‘Mykrobe predictor') that takes raw sequence data as input, and generates a clinician-friendly report within 3 minutes on a laptop. For S. aureus, the error rates of our method are comparable to gold-standard phenotypic methods, with sensitivity/specificity of 99.1%/99.6% across 12 antibiotics (using an independent validation set, n=470). For M. tuberculosis, our method predicts resistance with sensitivity/specificity of 82.6%/98.5% (independent validation set, n=1,609); sensitivity is lower here, probably because of limited understanding of the underlying genetic mechanisms. We give evidence that minor alleles improve detection of extremely drug-resistant strains, and demonstrate feasibility of the use of emerging single-molecule nanopore sequencing techniques for these purposes.
Abstract
 The clinical application of new sequencing techniques is expected to accelerate pathogen
identification. Here, Bradley
et al. present a clinician-friendly software package that uses sequencing data for quick
and accurate prediction of antibiotic resistance profiles for
S. aureus and
M. tuberculosis.
The clinical application of new sequencing techniques is expected to accelerate pathogen
identification. Here, Bradley
et al. present a clinician-friendly software package that uses sequencing data for quick
and accurate prediction of antibiotic resistance profiles for
S. aureus and
M. tuberculosis.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads.
- Record: found
- Abstract: found
- Article: not found
Transforming clinical microbiology with bacterial genome sequencing.
- Record: found
- Abstract: found
- Article: not found