- Record: found
- Abstract: found
- Article: found
BCL7A‐containing SWI/SNF/BAF complexes modulate mitochondrial bioenergetics during neural progenitor differentiation
Read this article at
Abstract
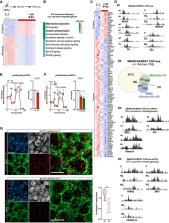
Mammalian SWI/SNF/BAF chromatin remodeling complexes influence cell lineage determination. While the contribution of these complexes to neural progenitor cell (NPC) proliferation and differentiation has been reported, little is known about the transcriptional profiles that determine neurogenesis or gliogenesis. Here, we report that BCL7A is a modulator of the SWI/SNF/BAF complex that stimulates the genome‐wide occupancy of the ATPase subunit BRG1. We demonstrate that BCL7A is dispensable for SWI/SNF/BAF complex integrity, whereas it is essential to regulate Notch/Wnt pathway signaling and mitochondrial bioenergetics in differentiating NPCs. Pharmacological stimulation of Wnt signaling restores mitochondrial respiration and attenuates the defective neurogenic patterns observed in NPCs lacking BCL7A. Consistently, treatment with an enhancer of mitochondrial biogenesis, pioglitazone, partially restores mitochondrial respiration and stimulates neuronal differentiation of BCL7A‐deficient NPCs. Using conditional BCL7A knockout mice, we reveal that BCL7A expression in NPCs and postmitotic neurons is required for neuronal plasticity and supports behavioral and cognitive performance. Together, our findings define the specific contribution of BCL7A‐containing SWI/SNF/BAF complexes to mitochondria‐driven NPC commitment, thereby providing a better understanding of the cell‐intrinsic transcriptional processes that connect metabolism, neuronal morphogenesis, and cognitive flexibility.
Abstract
Related collections
Most cited references105
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.

- Record: found
- Abstract: found
- Article: found
BEDTools: a flexible suite of utilities for comparing genomic features

- Record: found
- Abstract: found
- Article: found
