- Record: found
- Abstract: found
- Article: found
A large‐scale, multicenter characterization of BRAF G469V/A‐mutant non‐small cell lung cancer
Read this article at
Abstract
Background
Mutated BRAF is identified in 1%–5% non‐small cell lung cancer (NSCLC) patients, with non‐V600 mutations accounting for 50%–70% of these. The most common non‐V600 mutation is BRAF G469V/A. Currently, there are no targeted therapies available for non‐V600 mutated patients. A recent report provided interesting preclinical evidence revealing sensitivity of BRAF G469V to epidermal growth factor receptor (EGFR) inhibitors, raising the possibility of repurposing anti‐EGFR agents. It is therefore worthy to characterize the clinical and molecular features of BRAF G469V/A‐mutant NSCLC to provide more insights for precision therapy.
Methods
We conducted a retrospective screening of 25,694 Chinese patients with advanced or metastatic NSCLC to identify individuals with mutated BRAF. Additionally, we performed similar screenings on patients with adenocarcinoma (LUAD) from The Cancer Genome Atlas (TCGA) cohort ( n = 567) and the MSKCC cohort ( n = 1152). Subsequently, we characterized the clinical and molecular features of the patients carrying BRAF mutations.
Results
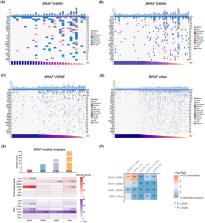
BRAF G469V was identified in 28 (0.1%) patients from the Chinese NSCLC cohort and 5 (0.9%) from TCGA‐LUAD. Notably, none was identified in the MSKCC cohort. G469A was found in 79 (0.3%) Chinese patients, 2 (0.4%) from TCGA‐LUAD, and 9 (0.8%) from the MSKCC cohort. Relative allele frequency analysis suggested most BRAF mutations as driven clones. Tumor mutation burden (median 4 mutations/Mb) was not significantly different between patients carrying G469V, G469A, V600E, or other BRAF mutations. Surprisingly, KRAS mutations were found in approximately 50% of patients with G469V mutation and about 8% of patients with G469A mutation, representing a prominent potential resistance mechanism against EGFR inhibitors. Structural modeling suggested BRAF G469V and G469A as binding partners of gefitinib.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: not found
Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries
- Record: found
- Abstract: found
- Article: not found
Tumor mutational load predicts survival after immunotherapy across multiple cancer types

- Record: found
- Abstract: found
- Article: found