- Record: found
- Abstract: found
- Article: found
Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
Read this article at
Abstract
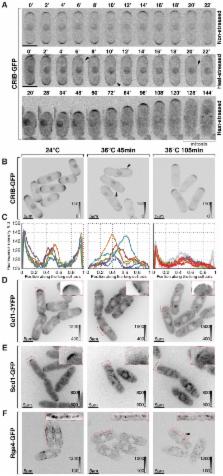
How the molecular mechanisms of stress response are integrated at the cellular level remains obscure. Here we show that the cellular polarity machinery in the fission yeast Schizosaccharomyces pombe undergoes dynamic adaptation to thermal stress resulting in a period of decreased Cdc42 activity and altered, monopolar growth. Cells where the heat stress-associated transcription was genetically upregulated exhibit similar growth patterning in the absence of temperature insults. We identify the Ssa2-Mas5/Hsp70-Hsp40 chaperone complex as repressor of the heat shock transcription factor Hsf1. Cells lacking this chaperone activity constitutively activate the heat-stress-associated transcriptional program. Interestingly, they also exhibit intermittent monopolar growth within a physiological temperature range and are unable to adapt to heat stress. We propose that by negatively regulating the heat stress-associated transcription, the Ssa2-Mas5 chaperone system could optimize cellular growth under different temperature regiments.
Author Summary
Heat stress, caused by fluctuations in ambient temperature, occurs frequently in nature. How organisms adapt and maintain regular patterns of growth over a range of environmental conditions remain poorly understood. Our work in the simple unicellular yeast Schizosaccharomyces pombe suggests that the heat stress-associated transcription must be repressed by the evolutionary conserved Hsp70-Hsp40 chaperone complex to allow cells to adapt the polarized growth machinery to elevated temperature.
Related collections
Most cited references91
- Record: found
- Abstract: found
- Article: not found
Cluster analysis and display of genome-wide expression patterns.

- Record: found
- Abstract: found
- Article: found
Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences
- Record: found
- Abstract: found
- Article: not found