- Record: found
- Abstract: found
- Article: found
A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase
Read this article at
Abstract
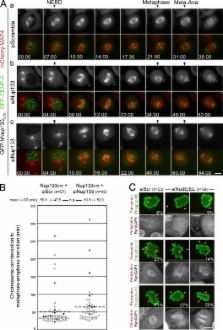
Nup133 links CENP-F, NudE/EL, and the dynein/dynactin complex to anchor centrosomes to the nuclear membrane.
Abstract
Centrosomes are closely associated with the nuclear envelope (NE) throughout the cell cycle and this association is maintained in prophase when they separate to establish the future mitotic spindle. At this stage, the kinetochore constituents CENP-F, NudE, NudEL, dynein, and dynactin accumulate at the NE. We demonstrate here that the N-terminal domain of the nuclear pore complex (NPC) protein Nup133, although largely dispensable for NPC assembly, is required for efficient anchoring of the dynein/dynactin complex to the NE in prophase. Nup133 exerts this function through an interaction network via CENP-F and NudE/EL. We show that this molecular chain is critical for maintaining centrosome association with the NE at mitotic entry and contributes to this process without interfering with the previously described RanBP2–BICD2-dependent pathway of centrosome anchoring. Finally, our study reveals that tethering of centrosomes to the nuclear surface at the G2/M transition contributes, along with other cellular mechanisms, to early stages of bipolar spindle assembly.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen.
- Record: found
- Abstract: found
- Article: not found
Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein).
- Record: found
- Abstract: found
- Article: not found