- Record: found
- Abstract: found
- Article: found
Elongating RNA polymerase II and RNA:DNA hybrids hinder fork progression and gene expression at sites of head-on replication-transcription collisions
Read this article at
Abstract
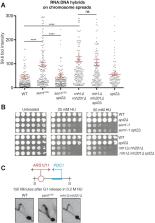
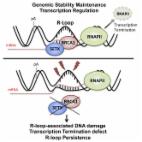
Uncoordinated clashes between replication forks and transcription cause replication stress and genome instability, which are hallmarks of cancer and neurodegeneration. Here, we investigate the outcomes of head-on replication-transcription collisions, using as a model system budding yeast mutants for the helicase Sen1, the ortholog of human Senataxin. We found that RNA Polymerase II accumulates together with RNA:DNA hybrids at sites of head-on collisions. The replication fork and RNA Polymerase II are both arrested during the clash, leading to DNA damage and, in the long run, the inhibition of gene expression. The inactivation of RNA Polymerase II elongation factors, such as the HMG-like protein Spt2 and the DISF and PAF complexes, but not alterations in chromatin structure, allows replication fork progression through transcribed regions. Attenuation of RNA Polymerase II elongation rescues RNA:DNA hybrid accumulation and DNA damage sensitivity caused by the absence of Sen1, but not of RNase H proteins, suggesting that such enzymes counteract toxic RNA:DNA hybrids at different stages of the cell cycle with Sen1 mainly acting in replication. We suggest that the main obstacle to replication fork progression is the elongating RNA Polymerase II engaged in an R-loop, rather than RNA:DNA hybrids per se or hybrid-associated chromatin modifications.
Graphical Abstract
Related collections
Most cited references98
- Record: found
- Abstract: found
- Article: not found
Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae.
- Record: found
- Abstract: found
- Article: found
