- Record: found
- Abstract: found
- Article: found
Manifold epigenetics: A conceptual model that guides engineering strategies to improve whole-body regenerative health
Read this article at
Abstract
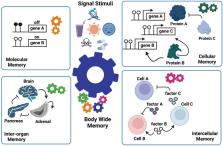
Despite the promising advances in regenerative medicine, there is a critical need for improved therapies. For example, delaying aging and improving healthspan is an imminent societal challenge. Our ability to identify biological cues as well as communications between cells and organs are keys to enhance regenerative health and improve patient care. Epigenetics represents one of the major biological mechanisms involving in tissue regeneration, and therefore can be viewed as a systemic (body-wide) control. However, how epigenetic regulations concertedly lead to the development of biological memories at the whole-body level remains unclear. Here, we review the evolving definitions of epigenetics and identify missing links. We then propose our Manifold Epigenetic Model (MEMo) as a conceptual framework to explain how epigenetic memory arises and discuss what strategies can be applied to manipulate the body-wide memory. In summary we provide a conceptual roadmap for the development of new engineering approaches to improve regenerative health.
Related collections
Most cited references101
- Record: found
- Abstract: found
- Article: found
The Hallmarks of Aging
- Record: found
- Abstract: found
- Article: not found
Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors.
- Record: found
- Abstract: found
- Article: not found