- Record: found
- Abstract: found
- Article: found
Blastocrithidia nonstop mitochondrial genome and its expression are remarkably insulated from nuclear codon reassignment

Read this article at
Abstract
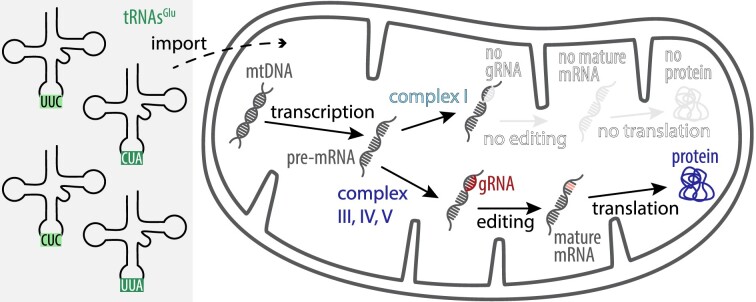
The canonical stop codons of the nuclear genome of the trypanosomatid Blastocrithidia nonstop are recoded. Here, we investigated the effect of this recoding on the mitochondrial genome and gene expression. Trypanosomatids possess a single mitochondrion and protein-coding transcripts of this genome require RNA editing in order to generate open reading frames of many transcripts encoded as ‘cryptogenes’. Small RNAs that can number in the hundreds direct editing and produce a mitochondrial transcriptome of unusual complexity. We find B. nonstop to have a typical trypanosomatid mitochondrial genetic code, which presumably requires the mitochondrion to disable utilization of the two nucleus-encoded suppressor tRNAs, which appear to be imported into the organelle. Alterations of the protein factors responsible for mRNA editing were also documented, but they have likely originated from sources other than B. nonstop nuclear genome recoding. The population of guide RNAs directing editing is minimal, yet virtually all genes for the plethora of known editing factors are still present. Most intriguingly, despite lacking complex I cryptogene guide RNAs, these cryptogene transcripts are stochastically edited to high levels.
Graphical Abstract
Related collections
Most cited references103

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found