- Record: found
- Abstract: found
- Article: found
ICTV Virus Taxonomy Profile: Mypoviridae 2023
review-article
Jens H. Kuhn
1 ,
Scott Adkins
2 ,
Katherine Brown
3 ,
Juan Carlos de la Torre
4 ,
Michele Digiaro
5 ,
Holly R. Hughes
6 ,
Sandra Junglen
7 ,
Amy J. Lambert
6 ,
Piet Maes
8 ,
Marco Marklewitz
9 ,
Gustavo Palacios
10 ,
Takahide Sasaya
11 ,
Massimo Turina
12 ,
Yong-Zhen Zhang
13
,
*
,
19 December 2023
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
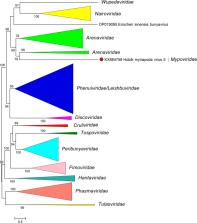
Mypoviridae is a family of negative-sense RNA viruses with genomes of about 16.0 kb that have been found in myriapods. The mypovirid genome consists of three monocistronic RNA segments that encode a nucleoprotein (NP), a glycoprotein (GP), and a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Mypoviridae, which is available at: ictv.global/report/mypoviridae.
Related collections
Most cited references5
- Record: found
- Abstract: found
- Article: not found
Redefining the invertebrate RNA virosphere
Mang Shi, Xian-Dan Lin, Jun-Hua Tian … (2017)

- Record: found
- Abstract: found
- Article: found
Re-assessing the diversity of negative strand RNA viruses in insects
Simon Käfer, Sofia Paraskevopoulou, Florian Zirkel … (2019)
- Record: found
- Abstract: not found
- Article: not found
RNA viromes from terrestrial sites across China expand environmental viral diversity
Yan-Mei Chen, Sabrina Sadiq, Jun-Hua Tian … (2022)