- Record: found
- Abstract: found
- Article: found
Interfering with retrotransposition by two types of CRISPR effectors: Cas12a and Cas13a
Read this article at
Abstract
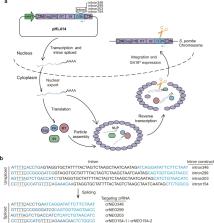
CRISPRs are a promising tool being explored in combating exogenous retroviral pathogens and in disabling endogenous retroviruses for organ transplantation. The Cas12a and Cas13a systems offer novel mechanisms of CRISPR actions that have not been evaluated for retrovirus interference. Particularly, a latest study revealed that the activated Cas13a provided bacterial hosts with a “passive protection” mechanism to defend against DNA phage infection by inducing cell growth arrest in infected cells, which is especially significant as it endows Cas13a, a RNA-targeting CRISPR effector, with mount defense against both RNA and DNA invaders. Here, by refitting long terminal repeat retrotransposon Tf1 as a model system, which shares common features with retrovirus regarding their replication mechanism and life cycle, we repurposed CRISPR-Cas12a and -Cas13a to interfere with Tf1 retrotransposition, and evaluated their different mechanisms of action. Cas12a exhibited strong inhibition on retrotransposition, allowing marginal Tf1 transposition that was likely the result of a lasting pool of Tf1 RNA/cDNA intermediates protected within virus-like particles. The residual activities, however, were completely eliminated with new constructs for persistent crRNA targeting. On the other hand, targeting Cas13a to Tf1 RNA intermediates significantly inhibited Tf1 retrotransposition. However, unlike in bacterial hosts, the sustained activation of Cas13a by Tf1 transcripts did not cause cell growth arrest in S. pombe, indicating that virus-activated Cas13a likely acted differently in eukaryotic cells. The study gained insight into the actions of novel CRISPR mechanisms in combating retroviral pathogens, and established system parameters for developing new strategies in treatment of retrovirus-related diseases.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells.
- Record: found
- Abstract: found
- Article: not found
Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells
- Record: found
- Abstract: found
- Article: not found