- Record: found
- Abstract: found
- Article: found
Genome-Wide Characterization of GRAS Family and Their Potential Roles in Cold Tolerance of Cucumber ( Cucumis sativus L.)
Read this article at
Abstract
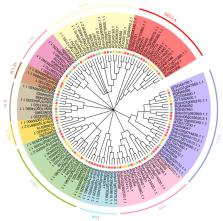
Cucumber ( Cucumis sativus L.) is one of the most important cucurbit vegetables but is often subjected to stress during cultivation. GRAS ( gibberellic acid insensitive, repressor of GAI, and scarecrow) genes encode a family of transcriptional factors that regulate plant growth and development. In the model plant Arabidopsis thaliana, GRAS family genes function in formation of axillary meristem and root radial structure, phytohormone (gibberellin) signal transduction, light signal transduction and abiotic/biological stress. In this study, a gene family was comprehensively analyzed from the aspects of evolutionary tree, gene structure, chromosome location, evolutionary and expression pattern by means of bioinformatics; 37 GRAS gene family members have been screened from cucumber. We reconstructed an evolutionary tree based on multiple sequence alignment of the typical GRAS domain and conserved motif sequences with those of other species ( A. thaliana and Solanum lycopersicum). Cucumber GRAS family was divided into 10 groups according to the classification of Arabidopsis and tomato genes. We conclude that tandem and segmental duplication have played important roles in the expansion and evolution of the cucumber GRAS ( CsaGRAS) family. Expression patterns of CsaGRAS genes in different tissues and under cold treatment, combined with gene ontology annotation and interaction network analysis, revealed potentially different functions for CsaGRAS genes in response to cold tolerance, with members of the SHR, SCR and DELLA subfamilies likely playing important roles. In conclusion, this study provides valuable information and candidate genes for improving cucumber tolerance to cold stress.
Related collections
Most cited references53
- Record: found
- Abstract: not found
- Book Chapter: not found
Protein Identification and Analysis Tools on the ExPASy Server

- Record: found
- Abstract: found
- Article: found
KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies
- Record: found
- Abstract: found
- Article: not found