- Record: found
- Abstract: found
- Article: found
Exit from Mitosis in Budding Yeast: Protein Phosphatase 1 is Required Downstream from Cdk1 Inactivation
Read this article at
Abstract
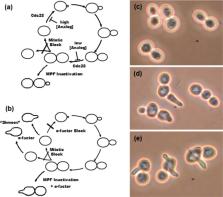
We show that inactivation of the protein kinase Cdk1/Cyclin B (Cdc28/Clb 2 in the budding yeast Saccharomyces cerevisiae) is not only necessary for cells to leave mitosis, as is well known, but also sufficient to trigger mitotic exit. Cells carrying the mutation cdc28- as1, which makes Cdc28 (Cdk1) uniquely sensitive to the ATP analog 1NM-PP1, were arrested with spindle poisons and then treated with 1NM-PP1 to inhibit Cdk1. This treatment caused the cells to exit mitosis and enter G1-phase as shown by initiation of rebudding (without cytokinesis), production of “shmoos” (when α-factor was present), stabilization of Sic1, and degradation of Clb2. This result provides a system in which to test whether particular gene products are required downstream from Cdk1 inactivation in exit from mitosis. In this system, the mutation cdc28- as1 is combined with a conditional mutation in the gene of interest. Using this approach, we demonstrate that Protein Phosphatase 1 (PPase1; Glc7 in S. cerevisiae) is required for reestablishment of G1-phase following Cdk1 inactivation. This system could be used to test whether other protein phosphatases are also needed downstream from Cdk1 inactivation, and it could be combined with phosphoproteomics to gain information about the substrates those phosphatases act on during mitotic exit.
Related collections
Most cited references102
- Record: found
- Abstract: found
- Article: not found
A chemical switch for inhibitor-sensitive alleles of any protein kinase.
- Record: found
- Abstract: found
- Article: not found
Global analysis of protein phosphorylation in yeast.
- Record: found
- Abstract: found
- Article: not found