- Record: found
- Abstract: found
- Article: found
Benchmarking Intrinsic Promoters and Terminators for Plant Synthetic Biology Research
Read this article at
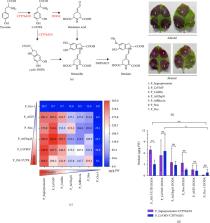
Abstract
The emerging plant synthetic metabolic engineering has been exhibiting great promise to produce either value-added metabolites or therapeutic proteins. However, promoters for plant pathway engineering are generally selected empirically. The quantitative characterization of plant-based promoters is essential for optimal control of gene expression in plant chassis. Here, we used N. benthamiana leaves and BY2 suspension cells to quantitatively characterize a library of plant promoters by transient expression of firefly/ Renilla luciferase. We validated the dual-luciferase reporter system by examining the correlation between reporter protein and mRNA levels. In addition, we investigated the effects of terminator–promoter combinations on gene expression and found that the combinations of promoters and terminators resulted in a 326-fold difference between the strongest and weakest performance, as reflected in reporter gene expression. As a proof of concept, we used the quantitatively characterized promoters to engineer the betalain pathway in N. benthamiana. Seven selected plant promoters with different expression strengths were used orthogonally to express CYP76AD1 and DODA, resulting in a final betalain production range of 6.0–362.4 μg/g fresh weight. Our systematic approach not only demonstrates the various intensities of multiple promoter sequences in N. benthamiana and BY2 cells but also adds to the toolbox of plant promoters for plant engineering.
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: found
- Article: not found
A golden gate modular cloning toolbox for plants.
- Record: found
- Abstract: found
- Article: not found