- Record: found
- Abstract: found
- Article: found
HMGA Proteins in Hematological Malignancies
Read this article at
Abstract
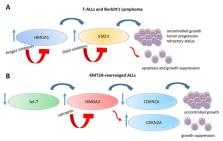
The high mobility group AT-Hook (HMGA) proteins are a family of nonhistone chromatin remodeling proteins known as “architectural transcriptional factors”. By binding the minor groove of AT-rich DNA sequences, they interact with the transcription apparatus, altering the chromatin modeling and regulating gene expression by either enhancing or suppressing the binding of the more usual transcriptional activators and repressors, although they do not themselves have any transcriptional activity. Their involvement in both benign and malignant neoplasias is well-known and supported by a large volume of studies. In this review, we focus on the role of the HMGA proteins in hematological malignancies, exploring the mechanisms through which they enhance neoplastic transformation and how this knowledge could be exploited to devise tailored therapeutic strategies.
Related collections
Most cited references202
- Record: found
- Abstract: found
- Article: not found
European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013.
- Record: found
- Abstract: found
- Article: not found
Selective blockade of microRNA processing by Lin28.
- Record: found
- Abstract: found
- Article: not found