- Record: found
- Abstract: found
- Article: found
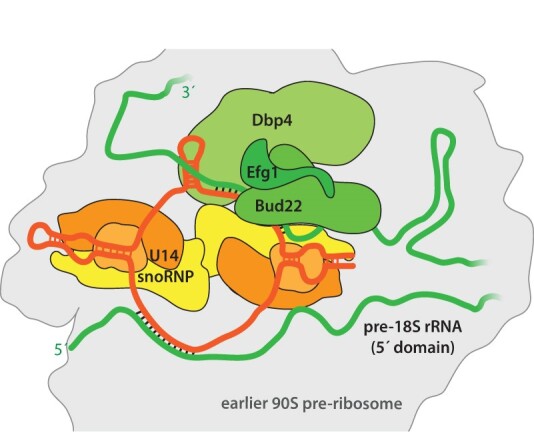
The Efg1–Bud22 dimer associates with the U14 snoRNP contacting the 5′ rRNA domain of an early 90S pre-ribosomal particle

Read this article at
Abstract
The DEAD-box helicase Dbp4 plays an essential role during the early assembly of the 40S ribosome, which is only poorly understood to date. By applying the yeast two-hybrid method and biochemical approaches, we discovered that Dbp4 interacts with the Efg1–Bud22 dimer. Both factors associate with early pre-90S particles and smaller complexes, each characterized by a high presence of the U14 snoRNA. A crosslink analysis of Bud22 revealed its contact to the U14 snoRNA and the 5′ domain of the nascent 18S rRNA, close to its U14 snoRNA hybridization site. Moreover, depletion of Bud22 or Efg1 specifically affects U14 snoRNA association with pre-ribosomal complexes. Accordingly, we concluded that the role of the Efg1–Bud22 dimer is linked to the U14 snoRNA function on early 90S ribosome intermediates chaperoning the 5′ domain of the nascent 18S rRNA. The successful rRNA folding of the 5′ domain and the release of Efg1, Bud22, Dpb4, U14 snoRNA and associated snoRNP factors allows the subsequent recruitment of the Kre33-Bfr2-Enp2-Lcp5 module towards the 90S pre-ribosome.
Graphical Abstract
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification.
- Record: found
- Abstract: found
- Article: not found
Gene Expression Omnibus: NCBI gene expression and hybridization array data repository.

- Record: found
- Abstract: found
- Article: found