- Record: found
- Abstract: found
- Article: found
Targeted 1H NMR metabolomics and immunological phenotyping of human fresh blood and serum samples discriminate between healthy individuals and inflammatory bowel disease patients treated with anti-TNF
Read this article at
Abstract
Abstract
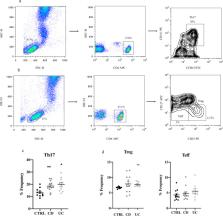
Inflammatory bowel disease is a multifactorial etiology, associated with environmental factors that can trigger both debut and relapses. A high level of tumor necrosis factor-α in the gut is the main consequence of immune system imbalance. The aim of treatment is to restore gut homeostasis. In this study, fresh blood and serum samples were used to identify biomarkers and to discriminate between Crohn’s disease and ulcerative colitis patients under remission treated with anti-TNF. Metabolomics based on Nuclear Magnetic Resonance spectroscopy (NMR) was used to detect unique biomarkers for each class of patients. Blood T lymphocyte repertories were characterized, as well as cytokine and transcription factor profiling, to complement the metabolomics data. Higher levels of homoserine-methionine and isobutyrate were identified as biomarkers of Crohn’s disease with ileocolic localization. For ulcerative colitis, lower levels of creatine-creatinine, proline, and tryptophan were found that reflect a deficit in the absorption of essential amino acids in the gut. T lymphocyte phenotyping and its functional profiling revealed that the overall inflammation was lower in Crohn’s disease patients than in those with ulcerative colitis. These results demonstrated that NMR metabolomics could be introduced as a high-throughput evaluation method in routine clinical practice to stratify both types of patients related to their pathology.
Key messages
-
NMR metabolomics is a non-invasive tool that could be implemented in the normal clinical practice for IBD to assess beneficial effect of the treatment.
-
NMR metabolomics is a useful tool for precision medicine, in order to sew a specific treatment to a specific group of patients.
-
Finding predictors of response to IFX would be desirable to select patients affected by IBD.
-
Immunological status of inflammations correlates with NMR metabolomics biomarkers.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts.

- Record: found
- Abstract: found
- Article: found
Role of intestinal microbiota and metabolites on gut homeostasis and human diseases
- Record: found
- Abstract: found
- Article: not found