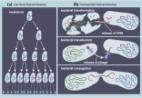
Antibiotics are now “endangered species” facing extinction due to the worldwide emergence of antibiotic resistance (ABR). Food animals are considered as key reservoirs of antibiotic-resistant bacteria with the use of antibiotics in the food production industry having contributed to the actual global challenge of ABR. There are no geographic boundaries to impede the worldwide spread of ABR. If preventive and containment measures are not applied locally, nationally and regionally, the limited interventions in one country, continent and for instance, in the developing world, could compromise the efficacy and endanger ABR containment policies implemented in other parts of the world, the best-managed high-resource countries included. Multifaceted, comprehensive, and integrated measures complying with the One Health approach are imperative to ensure food safety and security, effectively combat infectious diseases, curb the emergence and spread of ABR, and preserve the efficacy of antibiotics for future generations. Countries should follow the World Health Organization, World Organization for Animal Health, and the Food and Agriculture Organization of the United Nations recommendations to implement national action plans encompassing human, (food) animal, and environmental sectors to improve policies, interventions and activities that address the prevention and containment of ABR from farm-to-fork. This review covers (i) the origin of antibiotic resistance, (ii) pathways by which bacteria spread to humans from farm-to-fork, (iii) differences in levels of antibiotic resistance between developed and developing countries, and (iv) prevention and containment measures of antibiotic resistance in the food chain.