- Record: found
- Abstract: found
- Article: found
PLSDB: advancing a comprehensive database of bacterial plasmids
Read this article at
Abstract
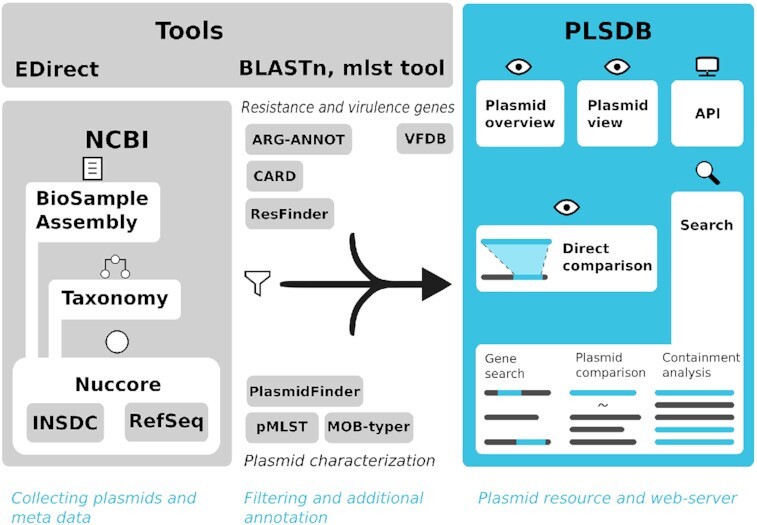
Plasmids are known to contain genes encoding for virulence factors and antibiotic resistance mechanisms. Their relevance in metagenomic data processing is steadily growing. However, with the increasing popularity and scale of metagenomics experiments, the number of reported plasmids is rapidly growing as well, amassing a considerable number of false positives due to undetected misassembles. Here, our previously published database PLSDB provides a reliable resource for researchers to quickly compare their sequences against selected and annotated previous findings. Within two years, the size of this resource has more than doubled from the initial 13,789 to now 34,513 entries over the course of eight regular data updates. For this update, we aggregated community feedback for major changes to the database featuring new analysis functionality as well as performance, quality, and accessibility improvements. New filtering steps, annotations, and preprocessing of existing records improve the quality of the provided data. Additionally, new features implemented in the web-server ease user interaction and allow for a deeper understanding of custom uploaded sequences, by visualizing similarity information. Lastly, an application programming interface was implemented along with a python library, to allow remote database queries in automated workflows. The latest release of PLSDB is freely accessible under https://www.ccb.uni-saarland.de/plsdb.
Graphical Abstract
Related collections
Most cited references37

- Record: found
- Abstract: found
- Article: found
BLAST+: architecture and applications

- Record: found
- Abstract: found
- Article: found
CD-HIT: accelerated for clustering the next-generation sequencing data

- Record: found
- Abstract: found
- Article: found