- Record: found
- Abstract: found
- Article: found
Subculturing and Gram staining of blood cultures flagged negative by the BACTEC™ FX system: Optimizing the workflow for detection of Cryptococcus neoformans in clinical specimens
research-article
Lingli Liu
1
,
2 ,
Lijun Du
1
,
3
,
4 ,
Shuquan He
1
,
5
,
6 ,
Tianshu Sun
7 ,
Fanrong Kong
8 ,
Yali Liu
1
,
2
,
9
,
*
,
,
Yingchun Xu
1
,
2
,
*
,
15 March 2023
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Objective
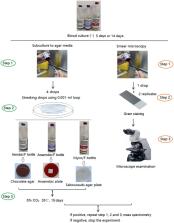
To investigate whether an incubation time of 5 days (Aerobic/F, Anaerobic/F) and 14 days (Myco/F) blood culture bottles is sufficient to prevent false-negative results.
Methods
We evaluated 1,244 blood bottles (344 patients) defined as negative by the BACTEC™ FX system. We also reviewed published cases and our own cases of bloodstream infection caused by Cryptococcus neoformans and simulated different scenarios, including different inoculation concentrations, bottle types, and clinical isolates.
Related collections
Most cited references37

- Record: found
- Abstract: found
- Article: found
fastp: an ultra-fast all-in-one FASTQ preprocessor
Shifu Chen, Yanqing Zhou, Yaru Chen … (2018)

- Record: found
- Abstract: found
- Article: found
Fast and accurate long-read alignment with Burrows–Wheeler transform
Heng Li, Richard Durbin (2010)

- Record: found
- Abstract: found
- Article: found
The Healthy Human Blood Microbiome: Fact or Fiction?
Diego J. Castillo, Riaan Rifkin, Don A. Cowan … (2019)