- Record: found
- Abstract: found
- Article: found
Long non-coding RNA-dependent mechanism to regulate heme biosynthesis and erythrocyte development
Read this article at
Abstract
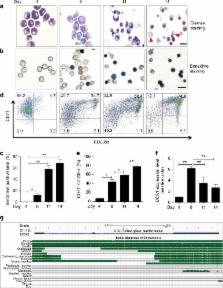
In addition to serving as a prosthetic group for enzymes and a hemoglobin structural component, heme is a crucial homeostatic regulator of erythroid cell development and function. While lncRNAs modulate diverse physiological and pathological cellular processes, their involvement in heme-dependent mechanisms is largely unexplored. In this study, we elucidated a lncRNA (UCA1)-mediated mechanism that regulates heme metabolism in human erythroid cells. We discovered that UCA1 expression is dynamically regulated during human erythroid maturation, with a maximal expression in proerythroblasts. UCA1 depletion predominantly impairs heme biosynthesis and arrests erythroid differentiation at the proerythroblast stage. Mechanistic analysis revealed that UCA1 physically interacts with the RNA-binding protein PTBP1, and UCA1 functions as an RNA scaffold to recruit PTBP1 to ALAS2 mRNA, which stabilizes ALAS2 mRNA. These results define a lncRNA-mediated posttranscriptional mechanism that provides a new dimension into how the fundamental heme biosynthetic process is regulated as a determinant of erythrocyte development.
Abstract
LncRNAs modulate diverse physiological cellular processes, however, their involvement in heme-dependent processes are not yet clear. Here the authors reveal the role of lncRNA UCA1 in erythroid cell development.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Braveheart, a long noncoding RNA required for cardiovascular lineage commitment.
- Record: found
- Abstract: found
- Article: not found
RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts.
- Record: found
- Abstract: found
- Article: not found