- Record: found
- Abstract: found
- Article: found
MmuPV1 E6 induces cell proliferation and other hallmarks of cancer
Read this article at
ABSTRACT
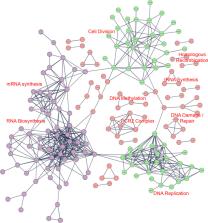
The E6 protein encoded by the murine papillomavirus (MmuPV1) is essential for MmuPV1-induced skin disease. Our previous work has identified a number of cellular interacting partners of MmuPV1 E6 and E7 through affinity purification/mass spectrometry analysis. These studies revealed that MmuPV1 E6 potently inhibits keratinocyte differentiation through multiple molecular mechanisms including inhibition of NOTCH and TGF-β signaling. Here, we report that MmuPV1 E6 has additional important oncogenic activities when expressed in its natural host cells, mouse keratinocytes, including increasing proliferation, overcoming density-mediated growth arrest, and proliferation under conditions of limited supply of growth factors. Unbiased proteomic/transcriptomic analyses of mouse keratinocytes expressing MmuPV1 E6 substantiated its effect on these cellular processes and divulged that some of these effects may be mediated in part through it upregulating E2F activity. Our analyses also revealed that MmuPV1 E6 may alter other cancer hallmarks including evasion of growth suppressors, inhibition of immune response, resistance to cell death, and alterations in DNA damage response. Collectively, our results suggest that MmuPV1 E6 is a major driver of multiple hallmarks of cancer in MmuPV1’s natural host cells, mouse keratinocytes.
IMPORTANCE
The Mus musculus papillomavirus 1 (MmuPV1) E6 and E7 proteins are required for MmuPV1-induced disease. Our understanding of the activities of MmuPV1 E6 has been based on affinity purification/mass spectrometry studies where cellular interacting partners of MmuPV1 E6 were identified, and these studies revealed that MmuPV1 E6 can inhibit keratinocyte differentiation through multiple mechanisms. We report that MmuPV1 E6 encodes additional activities including the induction of proliferation, resistance to density-mediated growth arrest, and decreased dependence on exogenous growth factors. Proteomic and transcriptomic analyses provided evidence that MmuPV1 E6 increases the expression and steady state levels of a number of cellular proteins that promote cellular proliferation and other hallmarks of cancer. These results indicate that MmuPV1 E6 is a major driver of MmuPV1-induced pathogenesis.
Related collections
Most cited references97
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: found