- Record: found
- Abstract: found
- Article: not found
Capturing Pluripotency
other
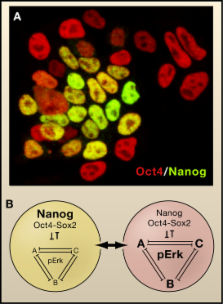
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
In this Essay, we argue that pluripotent epiblast founder cells in the embryo and embryonic stem (ES) cells in culture represent the ground state for a mammalian cell, signified by freedom from developmental specification or epigenetic restriction and capacity for autonomous self-replication. We speculate that cell-to-cell variation may be integral to the ES cell condition, safe-guarding self-renewal while continually presenting opportunities for lineage specification.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells.
Ian Chambers, Douglas Colby, Morag Robertson … (2003)
- Record: found
- Abstract: found
- Article: not found
BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3.
Qi Long Ying, Jennifer Nichols, Ian Chambers … (2003)
- Record: found
- Abstract: found
- Article: not found
Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway.
Claire Chazaud, Yojiro Yamanaka, Tony Pawson … (2006)
