- Record: found
- Abstract: found
- Article: found
Complete mitochondrial genome and the phylogenetic position of Halcyon smyrnensis (Aves: Coraciiformes)
research-article
Jiangyong Qu
a
,
b ,
Boyang Shi
a ,
Chenghua Guo
a ,
Jianhai Hou
a ,
Jiansen Cong
a ,
Jingrong Zhen
a
5 February 2016
2016
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
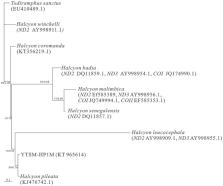
The complete mitochondrial genome of Halcyon smyrnensis was determined in this study. The mitogenomic length of Halcyon smyrnensis was 17 318 bp, including 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and one control region. Phylogenetic analyses show that H. leucocephala, H. pileata and H. smyrnensis congregated together, and our sample was sister to H. pileata.
Related collections
Most cited references6
- Record: found
- Abstract: found
- Article: not found
MRBAYES: Bayesian inference of phylogenetic trees.
J P Huelsenbeck, F Ronquist (2001)
- Record: found
- Abstract: found
- Article: not found
RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models.
Alexandros Stamatakis (2006)
- Record: found
- Abstract: not found
- Article: not found
Applications of mitochondrial DNA analysis in conservation: a critical review
C Möritz (1994)