- Record: found
- Abstract: found
- Article: found
A Single-Cell Transcriptome Atlas of the Human Pancreas
Read this article at
Summary
To understand organ function, it is important to have an inventory of its cell types and of their corresponding marker genes. This is a particularly challenging task for human tissues like the pancreas, because reliable markers are limited. Hence, transcriptome-wide studies are typically done on pooled islets of Langerhans, obscuring contributions from rare cell types and of potential subpopulations. To overcome this challenge, we developed an automated platform that uses FACS, robotics, and the CEL-Seq2 protocol to obtain the transcriptomes of thousands of single pancreatic cells from deceased organ donors, allowing in silico purification of all main pancreatic cell types. We identify cell type-specific transcription factors and a subpopulation of REG3A-positive acinar cells. We also show that CD24 and TM4SF4 expression can be used to sort live alpha and beta cells with high purity. This resource will be useful for developing a deeper understanding of pancreatic biology and pathophysiology of diabetes mellitus.
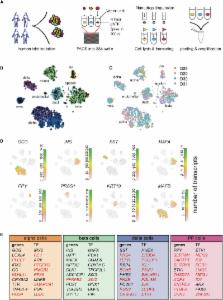
Graphical Abstract
Highlights
-
•
Single-cell sequencing of human pancreas allows in silico purification of cell types
-
•
We provide cell-type-specific genes, transcription factors, and cell-surface markers
-
•
StemID finds outlier populations of acinar and beta cells
-
•
CD24 and TM4SF4 function as two markers to enrich for alpha and beta cells
Abstract
Single-cell mRNA sequencing was used to describe the transcriptome of adult human pancreatic cell types. This resource was mined to find subpopulations of existing cell types and markers that can be used to purify live alpha and beta cells.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
The technology and biology of single-cell RNA sequencing.
- Record: found
- Abstract: found
- Article: not found
Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells
- Record: found
- Abstract: found
- Article: not found
Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.
