- Record: found
- Abstract: found
- Article: found
Genome-Wide Characterization of ISR Induced in Arabidopsis thaliana by Trichoderma hamatum T382 Against Botrytis cinerea Infection
Read this article at
Abstract
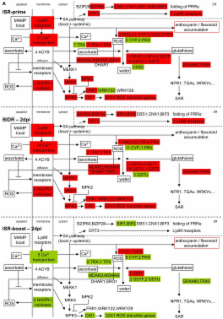
In this study, the molecular basis of the induced systemic resistance (ISR) in Arabidopsis thaliana by the biocontrol fungus Trichoderma hamatum T382 against the phytopathogen Botrytis cinerea B05-10 was unraveled by microarray analysis both before (ISR-prime) and after (ISR-boost) additional pathogen inoculation. The observed high numbers of differentially expressed genes allowed us to classify them according to the biological pathways in which they are involved. By focusing on pathways instead of genes, a holistic picture of the mechanisms underlying ISR emerged. In general, a close resemblance is observed between ISR-prime and systemic acquired resistance, the systemic defense response that is triggered in plants upon pathogen infection leading to increased resistance toward secondary infections. Treatment with T. hamatum T382 primes the plant (ISR-prime), resulting in an accelerated activation of the defense response against B. cinerea during ISR-boost and a subsequent moderation of the B. cinerea induced defense response. Microarray results were validated for representative genes by qRT-PCR. The involvement of various defense-related pathways was confirmed by phenotypic analysis of mutants affected in these pathways, thereby proving the validity of our approach. Combined with additional anthocyanin analysis data these results all point to the involvement of the phenylpropanoid pathway in T. hamatum T382-induced ISR.
Related collections
Most cited references109
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: found
In silico prediction of protein-protein interactions in human macrophages
- Record: found
- Abstract: found
- Article: not found