- Record: found
- Abstract: found
- Article: found
m 6A RNA methyltransferases METTL3/14 regulate immune responses to anti‐PD‐1 therapy
Read this article at
Abstract
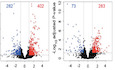
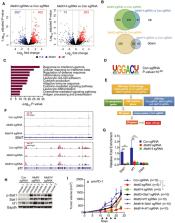
An impressive clinical success has been observed in treating a variety of cancers using immunotherapy with programmed cell death‐1 (PD‐1) checkpoint blockade. However, limited response in most patients treated with anti‐PD‐1 antibodies remains a challenge, requiring better understanding of molecular mechanisms limiting immunotherapy. In colorectal cancer (CRC) resistant to immunotherapy, mismatch‐repair‐proficient or microsatellite instability‐low (pMMR‐MSI‐L) tumors have low mutation burden and constitute ~85% of patients. Here, we show that inhibition of N 6 ‐methyladenosine (m 6A) mRNA modification by depletion of methyltransferases, Mettl3 and Mettl14, enhanced response to anti‐PD‐1 treatment in pMMR‐MSI‐L CRC and melanoma. Mettl3‐ or Mettl14‐deficient tumors increased cytotoxic tumor‐infiltrating CD8 + T cells and elevated secretion of IFN‐γ, Cxcl9, and Cxcl10 in tumor microenvironment in vivo. Mechanistically, Mettl3 or Mettl14 loss promoted IFN‐γ‐Stat1‐Irf1 signaling through stabilizing the Stat1 and Irf1 mRNA via Ythdf2. Finally, we found a negative correlation between METTL3 or METTL14 and STAT1 in 59 patients with pMMR‐MSI‐L CRC tumors. Altogether, our findings uncover a new awareness of the function of RNA methylation in adaptive immunity and provide METTL3 and METTL14 as potential therapeutic targets in anticancer immunotherapy.
Abstract
Related collections
Most cited references69
- Record: found
- Abstract: found
- Article: not found
Signatures of mutational processes in human cancer
- Record: found
- Abstract: found
- Article: not found
Cancer immunotherapy using checkpoint blockade
- Record: found
- Abstract: found
- Article: not found