- Record: found
- Abstract: found
- Article: found
Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in Southeast of Tanzania
Read this article at
Abstract
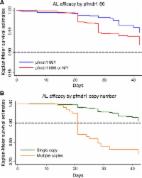
In Tanzania, chloroquine was replaced by sulphadoxine- pyrimethamine (SP) as a first-line for treatment of uncomplicated malaria. Due to high resistance in malaria parasites, SP lasted for only 5 years and by the end of 2006 it was replaced with the current artemisinin combination therapy. We therefore, set a study to determine the current genotypic mutations associated with Plasmodium falciparum resistance to artemisinin, partner drugs and chloroquine. Parasites DNA were extracted from dried blood spots collected by finger-prick from Tanzanian malaria infected patients. DNA were sequenced using MiSeq then genotypes were translated into drug resistance haplotypes at Wellcome Sanger Institute, UK. About 422 samples were successful sequenced for K13 gene (marker for artemisinin resistance), the wild type (WT) was found in 391 samples (92.7%) whereby 31 samples (7.3%) had mutations in K13 gene. Of 31 samples with mutations, one sample had R561H, a mutation that has been associated with delayed parasite clearance in Southeast Asia, another sample had A578S, a mutation not associated with artemisinin whilst 29 samples had K13 novel mutations. There were no mutations in PGB, EXO, P23_BP and PfMDR1 at position 86 and 1246 (markers for resistance in artemisinin partner drugs) but 270 samples (60.4%) had mutations at PfMDR1 Y184F. Additionally, genotyped PfCRT at positions 72–76 (major predictors for chroquine resistance), found WT gene in 443 out of 444 samples (99.8%). In conclusion, this study found mutations in K13-propeller gene and high prevalence of chloroquine susceptible P. falciparum in Southeast of Tanzania.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
A molecular marker of artemisinin-resistant Plasmodium falciparum malaria.
- Record: found
- Abstract: found
- Article: not found