- Record: found
- Abstract: found
- Article: found
Membrane curvature induces cardiolipin sorting
Read this article at
Abstract
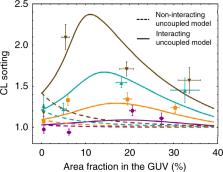
Cardiolipin is a cone-shaped lipid predominantly localized in curved membrane sites of bacteria and in the mitochondrial cristae. This specific localization has been argued to be geometry-driven, since the CL’s conical shape relaxes curvature frustration. Although previous evidence suggests a coupling between CL concentration and membrane shape in vivo, no precise experimental data are available for curvature-based CL sorting in vitro. Here, we test this hypothesis in experiments that isolate the effects of membrane curvature in lipid-bilayer nanotubes. CL sorting is observed with increasing tube curvature, reaching a maximum at optimal CL concentrations, a fact compatible with self-associative clustering. Observations are compatible with a model of membrane elasticity including van der Waals entropy, from which a negative intrinsic curvature of −1.1 nm −1 is predicted for CL. The results contribute to understanding the physicochemical interplay between membrane curvature and composition, providing key insights into mitochondrial and bacterial membrane organization and dynamics.
Abstract
Elena Beltrán-Heredia, Feng-Ching Tsai et al. examine the role of cardiolipin in membrane curvature, finding evidence compatible with a model based on membrane elasticity and van der Waals entropy. These results promote understanding of the interplay between membrane curvature and composition.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Aggregation and vesiculation of membrane proteins by curvature-mediated interactions.
- Record: found
- Abstract: found
- Article: not found
Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains.
- Record: found
- Abstract: found
- Article: not found